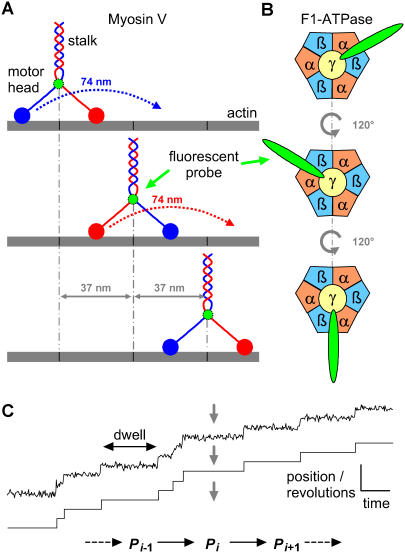
FIGURE 1.
Myosin V and F1-ATPase are typical molecular motors that convert chemical energy into mechanical energy by hydrolyzing ATP. (A) Myosin V is a dimeric motor protein, walking with a hand-over-hand mechanism along the actin filament, with the stalk taking 37-nm steps per ATP hydrolyzed (3). (B) F1-ATPase is a rotary motor that unidirectionally turns a rotor (γ-subunit) inside a stator (α3β3-complex). The rotor turns in 120° steps, one for each ATP hydrolyzed. Each 120° step consists of an 80° ATP-driven substep, followed by a 40° substep (8,9). (C) By attaching a probe to the motor protein, staircase data are constructed from single-molecule measurements. For a linear motor (myosin V), each dwell in the staircase data represents the location of the protein at a given position along its tracks, whereas for a rotary motor (F1-ATPase), each dwell corresponds to a given number of revolutions taken by the rotor. The duration of each dwell is random, with exponential distribution determined by kinetics and by experimental conditions, such as ATP concentration or applied mechanical force. Due to finite sampling time, more than one step may occur within the sampling interval, resulting in missed events. At each position, the motor undergoes transitions between two or more conformations. One of these transitions is ATP binding.