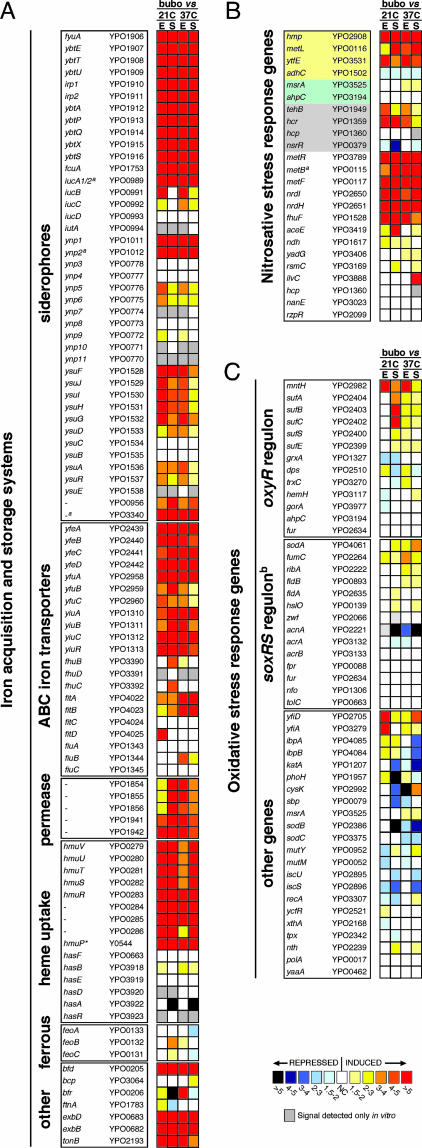
Fig. 2.
Differential expression in the bubo of genes implicated in the response to iron limitation (A) (9), nitrosative stress (B) (13–20), and oxidative stress (C) (22, 39). The mean fold increase or decrease in individual gene expression in the bubo compared with exponential (E) and stationary (S) phase cultures at 21°C and 37°C is indicated by the color scale bar. For the nitrosative stress gene list (B), genes specifically required for resistance to NO and GSNO or to peroxynitrite are highlighted in yellow and green, respectively. Genes highlighted in gray, along with hmp and ytfE, are predicted to be up-regulated in response to RNS by repression of the NO-sensitive negative regulator encoded by nsrR (17, 18); the other genes were shown to be up-regulated at least 2-fold in response to NO or GSNO stress in three different in vitro microarray studies (13–15). The YPO and Y numbers refer to the Y. pestis CO92 and KIM genome annotations, respectively (10, 11). ∗, not annotated in Y. pestis CO92; a, pseudogene; b, the Y. pestis response to superoxide may not occur via SoxRS regulation because no soxRS homologs were identified in the genome (10, 11).