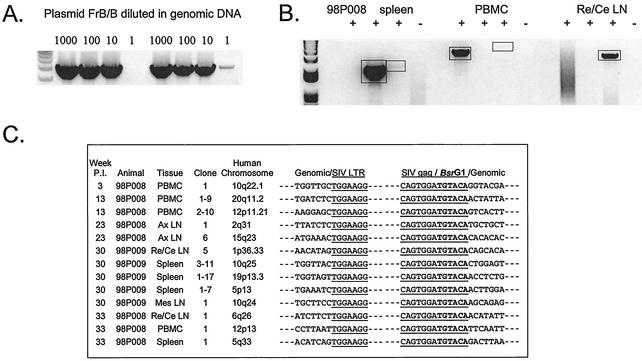
FIG. 5.
Detection of integrated SIV DNA in resting CD4+ T lymphocytes by inverse PCR. (A) Sensitivity of inverse PCR. The plasmid FrB/B was constructed by excising a BamHI/BlpI fragment between the middle of the gag gene and the end of 3′ LTR region from a plasmid containing the SIV/17E-Fr provirus. Inverse PCR amplification of this template gives a 4.5-kb product. This plasmid was serially diluted into 200 ng of genomic DNA as a control for the initial and nested PCRs. Results of two independent PCRs are shown. (B) Representative inverse PCRs from different tissues from treated animal 98P008. Lanes indicate the addition (+) and omission (−) of ligase in the ligation reaction, respectively. Bands were cloned and sequenced. Re/Ce LN, retropharyngeal and cervical lymph nodes. (C) Inverse PCR allows definitive identification of integration sites. Integration sites are shown as sequences immediately upstream of the LTR. Note that the LTR shows the characteristic loss of the terminal two nucleotides (5′AC) resulting from the cleavage by integrase of the two terminal bases from the 3′ end of each strand of the full-length linear SIV genome. The other junction results from ligation of cut BsrG1 sites in gag and in macaque DNA upstream of the integration site. The macaque sequences cloned by this method are each homologous (>90%) to the indicated sequences in the human genome. Ax LN, axillary lymph nodes; Mes LN, mesenteric lymph nodes.