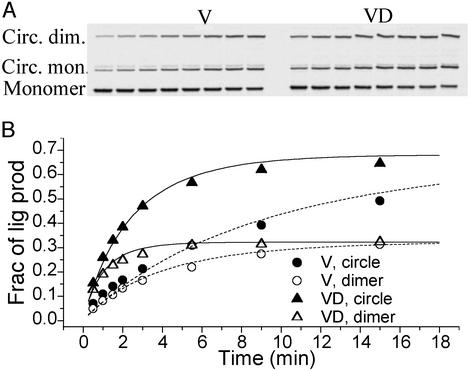
Figure 5.
The cyclization kinetics of the 126-bp unlabeled (V) and dye-labeled (VD) DNA constructs with CGAT cohesive ends. The gel autoradiography is shown in A and the quantitative evolution of each ligation product is presented in B. Both cyclizations were carried out with the same 20 nM total DNA and 160 units/ml T4 ligase. The best-fit parameters for V are: k1 = 0.082 min−1, k2 = 0.0022 min−1⋅nM−1, J = 36.7 nM; for VD: k1 = 0.31 min−1, k2 = 0.0084 min−1⋅nM−1, J = 37.2 nM. The dashed and solid curves for V and VD, respectively, were calculated with the same formula as in ref. 11, except for the symmetric factor from the palindromic nature of the cohesive ends. The weak bands slightly above those corresponding to circular monomer barely change with time and were not identified. The ligatable fractions of both V and VD molecules were estimated to be ≈80%, and only these fractions were analyzed in B.