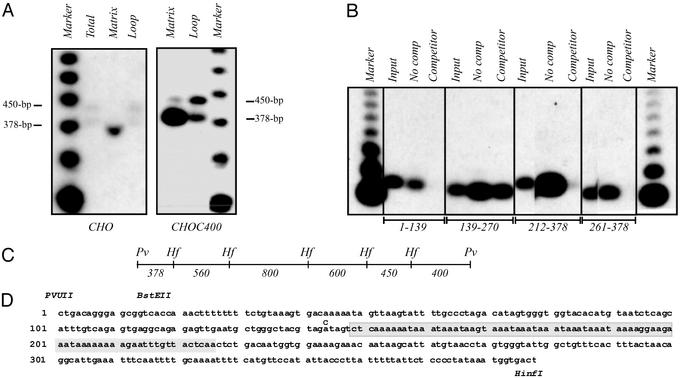
Figure 2.
Defining the minimal MAR-binding sequence. (A) Matrix-halo structures were prepared from CHO and CHOC400 cells as described in Materials and Methods, and loop DNA was removed with a combination of PvuII and HinfI. Equal weights of matrix and loop DNA fractions were separated on an agarose gel and blotted to a nylon membrane. The transfer was then hybridized with a combination of 450- and 379-bp PvuII/HinfI subfragments. The marker is an end-labeled 123-bp ladder (BRL). (B) Matrix/halo structures were prepared from CHOC400 cells and all attached DNA was completely removed with DNaseI. Isolated DNA-free matrices were then incubated with end-labeled fragments from the 3.4-kb PvuII MAR-containing fragment in the presence and absence of cold competitor DNA. The bound radioactive DNA was analyzed by Southern blotting. (C) HinfI subfragments of the 3.4-kb PvuII MAR-binding fragment (22), which were tested for association with the matrix in the in vivo assay illustrated in A (additional probings not shown). (D) Primary sequence of the 378-bp PvuII/HinfI MAR-binding fragment. Relevant restriction enzyme sites are shown above the sequence, and the 78-bp AT-rich deletion in the AT-MARKO cell line is shaded. At position 144, a C was inserted in the donor BAC that gave rise to the AT-MARKO cell line to create an SpeI site for diagnosis of recombinants. The sequence shown is from PCR products obtained from DR8A-7, as well as a subclone from a CHOC400 cosmid library. Sequence was also obtained from the AT-MARKO cell line to confirm deletion of the shaded region.