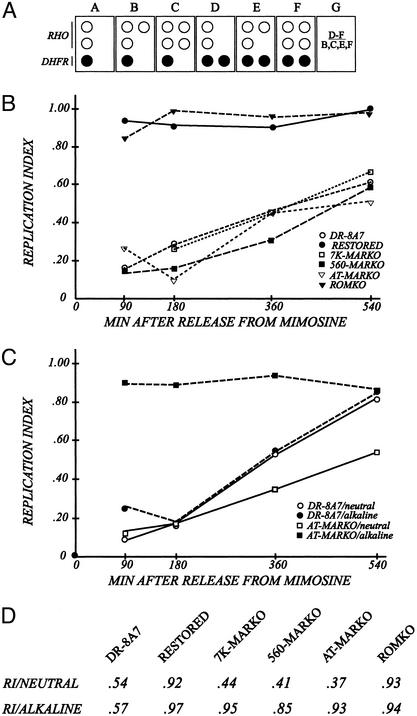
Figure 3.
The MARKO deletion variants appear to be very late replicating by the criterion of the FISH-based replication timing assay. (A) Principle of the FISH-based replication timing assay (37). Cells are swollen spread on microscope slides (34, 40), and hybridized with a digoxygenin-labeled cosmid specific for the intergenic region (KD504) and with a biotin-labeled cosmid specific for the control, early-replicating, rhodopsin origin. The respective signals are detected with fluorescein-labeled antidigoxygenin or Texas red-labeled antiavidin, as outlined in Materials and Methods (40). The number of dots of each color is recorded for at least 100 interphase nuclei from each cell line. Boxes B–F represent cells in different stages of the S-period or G2. Box G defines a replication index, which is calculated by dividing the number of cells that have replicated the DHFR locus (as in boxes D–F) by the number that has doubled the rhodopsin control locus (as in boxes B, C, E, and F). (B) Cells were sampled from synchronized cultures at the indicated times and swollen under neutral conditions. Resulting replication indices for each cell line are plotted as a function of time after release from the G1/S mimosine block. (C) Synchronized AT-MARKO and DR8-A7 cell lines were harvested at the indicated times, divided into two, and prepared for the FISH assay under either neutral or alkaline conditions. After hybridization with the mixed DHFR/rhodopsin probe, replication indices were determined. (D) Unsynchronized cultures of each of the indicated cell lines were divided into two and prepared for FISH under either neutral or alkaline swelling conditions. Replication indices were determined as described above.