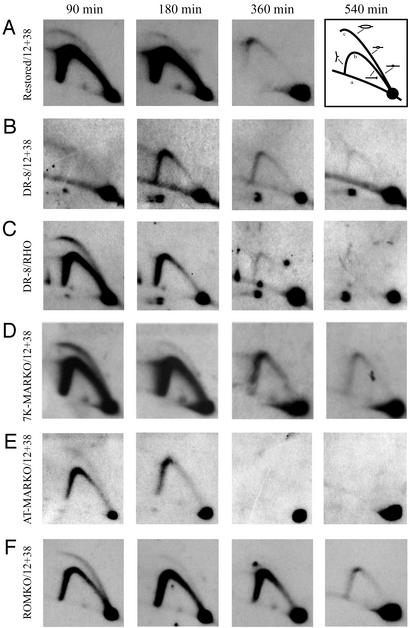
Figure 4.
2D gel analysis of the MARKO variants suggests that deletion of the MAR has no effect on either the efficiency or timing of initiation in the DHFR locus. Replication intermediates were purified from aliquots of the same synchronized cell populations analyzed by the FISH-based assay in Fig. 3B. After digestion with EcoRI, intermediates were separated on a neutral/neutral 2D gel, transferred to Hybond N+, and hybridized with a combination of probes 12 and 38, which are specific for a fragment containing ori-β. Each cell line was additionally analyzed with a probe specific for the early-replicating rhodopsin standard (shown here for the DR8-A7 variant only). The principle of the method is outlined in A Right. Replication intermediates are separated in the first dimension according to molecular mass, which for any fragment will vary from 1n (unreplicated) to just less than 2n. The first dimension lane is excised, turned through 90°, and separated in the second dimension according to both mass and shape (32). Linear nonreplicating fragments trace a diagonal (curve a). If a fragment is replicated passively by a fork originating from a site outside of the fragment, it will display a single fork arc (curve b). However, if the fragment contains a centered initiation site, it will display an arching bubble arc that extends from the 1n to the 2n positions (curve c).