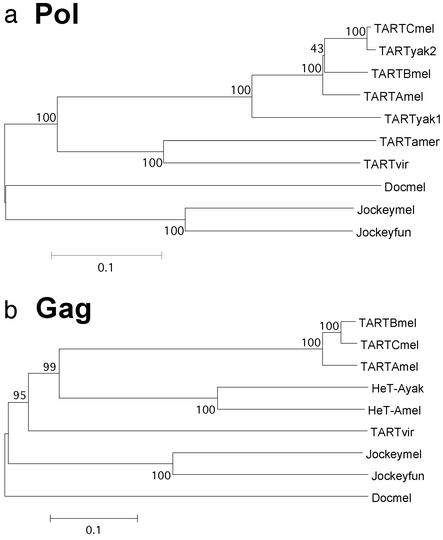
Figure 5.
Phylogenetic relationships of TART, HeT-A, Doc, and jockey elements in different Drosophila species. Neighbor-joining trees for the protein sequences are shown. (The UPGMA trees yield the same relationships as do the nucleotide trees.) Bootstrap tests were performed with 500 replications and a cut-off value of 50% for the consensus tree. Numbers indicate bootstrap values of >40% in the corresponding node. Scale bar corresponds to the P value (number of differences normalized by the number of total residues). Elements are indicted by the first three letters of the species (fun, Drosophila funebris). Note that TARTyak2 groups with TARTmelC as proposed (7). Gag from TARTyak was not included because all of the fragments cloned to date are 5′ truncated. GenBank accession numbers: jockeymel ORF1, M22874; ORF2, AAA28675; jockeyfun, PIR:B38418; Doc, CAA35587.