Figure 1.
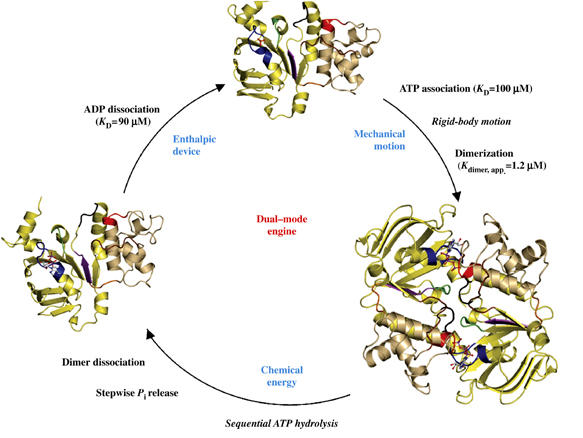
The catalytic cycle of the HlyB-NBD. Crystal structures of the monomeric nucleotide-free (Schmitt et al, 2003), dimeric ATP-bound (H662A and E631Q) and monomeric ADP-bound (wild type, E631Q and H662A) forms (this study) are shown. For simplicity, the structure of the ATP/Mg2+-bound form (Zaitseva et al, 2005a) is not shown. The catalytic domain is colored in light yellow and the helical domain in light tan. Conserved motifs are highlighted and color-coded as follows: Walker A (residues 502–510, blue), Q-loop (residues 549–556, brown), ABC-signature motif (residues 606–610, red), Pro-loop (residues 623–625, orange), Walker B (residues 626–630, magenta), D-loop (residues 634–637, black) and H-loop (residues 661–663, green). Bound ligands are shown in ball-and-stick representation. KD values were taken from Zaitseva et al (2005b).
