Abstract
The DnaE intein in Synechocystis sp. strain PCC6803 is the first and only naturally split intein that has been identified so far. It is capable of catalyzing a protein trans-splicing mechanism to assemble a mature protein from two separate precursors. Therefore, it is a powerful tool for protein modification and engineering. Inteins have not been identified, nor have intein-mediated protein splicing reactions been demonstrated, in plant cells. In this paper, we describe the use of the Ssp DnaE split intein in transgenic plants for reconstitution of a protein trans-splicing reaction. We have synthesized artificial genes that encode for N-terminal half (Int-n) and C-terminal half (Int-c) fragments of Ssp DnaE split intein and divided β-glucuronidase (GUS) gene to encode GUS-n and GUS-c parts of the enzyme as reporter. The in-frame fusions of GUSn/Intn and Intc/GUSc were constructed and transfected into Arabidopsis. We have observed in vivo reassembly of functional β-glucuronidase when both GUSn/Intn and Intc/GUSc constructs were introduced into the same Arabidopsis genome either by cotransformation or through genetic crossing, hereby signifying an intein-mediated protein trans-splicing mechanism reconstituted in plant cells.
Inteins are internal protein elements mediating posttranslational protein splicing. During this process, the intein element in a protein precursor catalyzes a series of reactions to remove itself from the precursor and ligate the flanking external protein fragments (i.e., exteins) into a mature protein (1). A typical intein element consists of 400–500 amino acid residues. It contains four conserved protein splicing motifs, A, B, F, and G, as well as a homing endonuclease sequence embedded between motifs B and F (1, 2). The endonuclease plays a role in mobilizing intein genes. It can be deleted from the intein sequence without compromising protein splicing (3, 4). In fact, naturally occurring mini-inteins have been identified from various organisms according to InBase, the intein database (5). Three conserved amino acid residues at intein–extein junctions are directly involved in the protein splicing reactions. They include a Ser or Cys at the intein N terminus (the first amino acid in motif A), an Asn or Gln at the intein C terminus (the last amino acid in motif G), and a Ser, Thr, or Cys at the beginning of the C-extein. The protein splicing mechanism consists of four coupled nucleophilic displacements: (i) an N-O or N-S acyl shift at the N-terminal splice junction that replaces the peptide bond with an ester or thioester bond; (ii) a transesterification reaction resulting in cleavage at the N-terminal splice junction and transfer of the N-extein to the side chain of the first amino acid in the C-extein; (iii) cyclization of the intein C-terminal Asn or Gln residue to release the intein; and (iv) an O-N or S-N acyl shift to form a peptide bond between the exteins (1, 2). Because a classic protein splicing reaction ligates two parts of the same protein precursor, it is termed protein cis-splicing.
Inteins can be split into an N-terminal half (Int-n) that contains the A and B motifs and a C-terminal half (Int-c) that contains the F and G motifs. When the split intein fragments are fused with different proteins or peptides, they are able to conduct protein trans-splicing that assembles two separate precursor molecules into a mature hybrid molecule both in vitro and in vivo (6–9). Recently, a pair of functional and naturally split intein-coding sequences were identified from the split DnaE genes in the genome of Synechocystis sp. PCC6803 (10). One of the split genes encodes a fusion protein tandem containing 774 N-terminal amino acid residues of the DnaE protein and 123 N-terminal amino acid residues of the DnaE intein. The other split gene encodes a protein sequence for the 36 C-terminal amino acid residues of the DnaE intein, followed by the 423 C-terminal aa of the DnaE protein. Although the two genes are located 745-kb apart on opposite strands of the Ssp PCC6803 genome, the mature protein product is an intact 1,197-aa catalytic subunit of DNA polymerase III, lacking any intein sequence because of the protein trans-splicing. The Ssp DnaE intein can mediate a trans-splicing reaction when fused to foreign proteins, as demonstrated by the reassembly of functional enzymes in Escherichia coli (11, 12). In a special case of the trans-splicing, it also can catalyze the head-to-tail cyclization of a target protein if the precursor has a structure of Int-c∷Target∷Int-n (13). Compared to artificially split inteins, the DnaE intein catalyzes protein trans-splicing reactions with higher efficiency and under milder conditions (12).
Intein-mediated protein trans-splicing has been introduced into mammalian cells for various applications, such as detecting protein–protein interactions in Chinese hamster ovary cells (14) and generating cyclic peptide libraries in human B cells (15). This technology is also an attractive tool for plant genetic engineering. By using an endogenous or exogenous protein splicing mechanism in plant cells, unique protein assembly machinery could be introduced to produce and modify complex protein polymers or to function as a sensitive molecular switch. Recent work on Ssp DnaE split intein has suggested that this intein could be used in developing and applying herbicide-resistant crops in an environmentally friendly way (11). However, among nearly 140 putative inteins registered in InBase, none has been found from higher plants (5). Nor has any known intein been demonstrated to function in higher plants. This lack of evidence presents reasonable doubt as to whether inteins will function in plant cells. In this report, we use the DnaE split intein to reassemble the divided fragments of β-glucuronidase (GUS) in transgenic Arabidopsis. Our experiments show that the DnaE split intein can catalyze β-glucuronidase reassembly, thus demonstrating a potential utility for intein application in plants.
Materials and Methods
Gene Fusion and Plasmid Construction.
Oligonucleotides, shown as the first four groups in Table 1, were synthesized to make plant-optimized Ssp DnaE Int-n- and Int-c-coding sequences. After phosphorylation by using ATP and polynucleotide kinase, all of the oligonucleotides in groups 1 and 2 were mixed and annealed to assemble Pint-n. Those in groups 3 and 4 were used to assemble Pint-c. The annealing was carried out by heating the mixtures at 98°C for 10 min and then cooling to 23°C at 1°C per min. The annealed oligonucleotides were then ligated, and the correctly assembled fragments were amplified from the mixtures by PCR reaction by using Pfu DNA polymerase (Stratagene). Finally, Pint-n and Pint-c were subcloned into PCR-Script Amp (Stratagene), and a DNA fragment containing a 2-μm yeast replication origin and a Trp selective marker (16, 17) was integrated into each clone, resulting in plasmids pPint-n and pPint-c.
Table 1.
A summary of oligonucleotides for Pint-n and Pint-c synthesis
| Oligo | Sequences (5′–3′) |
|---|---|
| Group 1 | |
| IntN+1 | TGCCTTTCTTTCGGAACTGAGATCCTTACCGTTGAGTACGGACCACTTCCTATTGGTAAGATCGTTTCTGAGGAA |
| IntN+2 | ATTAACTGCTCAGTGTACTCTGTTGATCCAGAAGGAAGAGTTTACACTCAGGCTATCGCACAATGGCACGATAGG |
| IntN+3 | GGTGAACAAGAGGTTCTCGAGTACGAGCTTGAAGATGGATCCGTTATTCGTGCTACCTCTGACCATAGATTCTTG |
| IntN+4 | ACTACAGATTATCAGCTTCTCGCTATCGAGGAAATCTTTGCTAGGCAACTTGATCTCCTTACTTTGGAGAACATC |
| IntN+5 | AAGCAGACAGAAGAGGCTCTTGACAACCACAGACTTCCATTCCCTTTGCTCGATGCTGGAACCATCAAG |
| Group 2 | |
| IntN−1 | CTTGATGGTTCCAGCATCGAGCAAAGGGAA |
| IntN−2 | TGGAAGTCTGTGGTTGTCAAGAGCCTCTTCTGTCTGCTTGATGTTCTCCAAAGTAAGGAGATCAAGTTGCCTAGC |
| IntN−3 | AAAGATTTCCTCGATAGCGAGAAGCTGATAATCTGTAGTCAAGAATCTATGGTCAGAGGTAGCACGAATAACGGA |
| IntN−4 | TCCATCTTCAAGCTCGTACTCGAGAACCTCTTGTTCACCCCTATCGTGCCATTGTGCGATAGCCTGAGTGTAAAC |
| IntN−5 | TCTTCCTTCTGGATCAACAGAGTACACTGAGCAGTTAATTTCCTCAGAAACGATCTTACCAATAGGAAGTGGTCC |
| IntN−6 | GTACTCAACGGTAAGGATCTCAGTTCCGAAAGAAAGGCA |
| Group 3 | |
| IntC+1 | ATGGTTAAGGTGATTGGAAGACGTTCTCTTGGTGTTCAAAGGATCTTCGATATCGGATTGCCACAAGACCACAAC |
| IntC+2 | TTTCTTCTCGCTAATGGTGCCATCGCTGCCAATTGC |
| Group 4 | |
| IntC−1 | GCAATTGGCAGCGATGGCACCATTAGCGAGAAGAAAGTTGTGGTCTTGTGGCAATCCGATATCGAAGATCCTTTG |
| IntC−2 | AACACCAAGAGAACGTCTTCCAATCACCTTAACCAT |
| Group 5 | |
| BSGusN(+) | CCCCTCGAGGTCGACGGTATCGATATCCATGGCTCATCATCATCA |
| IntNGusN(−) | GGATCTCAGTTCCGAAAGAAAGGCAGTCTTGCGCGACATGCGTCA |
| IntN(6)GusN(−) | GTCCGTACTCAACGGTAAGGATCTCGTCTTGCGCGACATGCGTCA |
| IntCGusC(+) | CGCTAATGGTGCCATCGCTGCCAATTGTAACCACGCGTCTGTTGA |
| BS(−) | CGAGGTCGACGGTATCGATAAG |
The GUS gene (18) in a pBluescript SK(+) clone was modified by adding 6xHis tag-coding sequences at its 5′- and 3′-ends. The modification yielded plasmid pHGUSH. To make the in-frame fusions between β-glucuronidase and DnaE split intein, the modified GUS gene was divided and integrated into Pint-n and Pint-c by yeast-based PCR-directed DNA recombination (16, 17). For this purpose, the five oligonucleotides of group 5 were synthesized (Table 1). GUS fragments GUS-n, GUS-n(6), and GUS-c were amplified from pHGUSH by PCR, using oligonucleotide pairs of BSGusN(+)/IntNGusN(−), BSGusN(+)/IntN(6)GusN(−), and IntCGusC(+)/BS(−), respectively. Because these fragments also included 25-nt sequences at their 5′- and 3′-ends, that are complementary to the target sequences in pPint-n or pPint-c, they can be accurately integrated into pPint-n and pPint-c to create coding sequences for the designed in-frame fusions.
To construct expression cassettes, the fusion protein-coding sequences were isolated from the plasmids derived from the PCR-directed recombination and inserted between the cauliflower mosaic virus 35S promoter and nopaline synthase terminator. Finally, these cassettes were isolated and inserted into the T-DNA region in the pZBL1 binary vector (19) either in combination or individually, resulting in the designated expression plasmids.
Transformation and Genetic Crossing of Arabidopsis.
The expression plasmids were introduced into Agrobacterium strain C58C1(pMP90) and transformed into Arabidopsis thaliana (ecotype Columbia) by an Agrobacterium-mediated floral dip transformation method (20). The treated plants were grown to maturation under standard conditions, and a mixture of nontransformed and primary transformed seeds (T1) was collected. Primary transformants were identified by germinating T1 seeds on selective medium (20) containing 50 mg/liter kanamycin sulfate (Sigma) to select for the neomycin phosphotransferase (NPTII) marker or 20 mg/liter glufosinate ammonium (Riedel-de Haën, Seelze, Germany) to select for the bialaphos resistance (Bar) marker. Individual plants were then grown in soil to produce T2 seeds to represent individual transformation events.
To select homozygous plants, T2 seeds were germinated on the selective medium again. Numbers of green and healthy or pale and dying seedlings for each transformation event were subjected to a χ-squared test (21) to identify plants with a single transgene insertion (indicated as a typical 3:1 segregation ratio). Thus, eight T2 seedlings from each of those single-insertion events were further grown in soil. T3 seeds were collected from individual plants and germinated on appropriate selective medium. Homozygous plants were identified if T3 seeds could germinate without segregation.
For crosses, pollen was collected from fully opened flowers of homozygous pollen donor plants and dusted onto the stigmas of the homozygous emasculated recipient plants prepared from unopened buds. Hybrids were confirmed by germinating progeny of the crosses on selective media and were further grown in soil until maturation. Seeds from each hybrid plant were collected separately to represent individual primary hybrids.
Assays for Transgene Expression and Protein Splicing Reconstitution.
Activity of β-glucuronidase in the transgenic plants was determined by a GUS histochemical-staining assay (22).
For RNA-filter hybridization assay, total RNA was prepared from plant tissues by using RNeasy Plant Mini kit (Qiagen). The RNA concentration was determined with spectral absorption at 260 nm. RNA (≈6 μg) was fractionated in an agarose gel by electrophoresis, transferred to nylon membrane, and probed with 32P-α-dCTP-labeled GUS-coding sequence, following standard protocols (23).
To carry out protein immunoblot assays, total soluble protein was extracted by grinding plant tissues in 2× volume of protein extract buffer (50 mM Tris⋅HCl, pH 7.5/50 mM NaCl/0.1 mM EDTA/5 mM MgCl2/5% glycerol/1% Sigma proteinase inhibitor mixture). Protein concentration was determined by using the Bio-Rad Protein Assay reagent. Total soluble protein (15 μg) was subjected to the assay, following standard protocols (24). Penta-His Ab (Qiagen) was used to detect both N- and C-terminal 6xHis tags, whereas anti-His(C Term)-horseradish peroxidase (HRP) Ab (Invitrogen) for C-terminal 6xHis tag only. When the blot needed to be reprobed, the Ab in the previous assay was stripped from the membrane in a solution containing 0.5 M NaCl and 0.2 M glycine (pH 2.8).
Results
Plant-Optimized DnaE-Intein-Coding Sequences and GUS–Intein Fusions.
To use Ssp DnaE split intein in transgenic plants, plant-optimized intein-coding sequences were synthesized and assembled de novo based on the peptide sequences of the DnaE split intein in Synechocystis sp. PCC6803 (10) and the rules of codon usage in plants (25). The oligonucleotides in groups 1 and 2 were assembled into a double-strand DNA fragment Pint-n (GenBank accession no. AF545505), that encoded for Ssp DnaE Int-n. Similarly, the oligonucleotides in groups 3 and 4 were assembled into a double-strand DNA fragment Pint-c (GenBank accession no. AF545504) to encode Ssp DnaE Int-c and an additional C-terminal Cys codon.
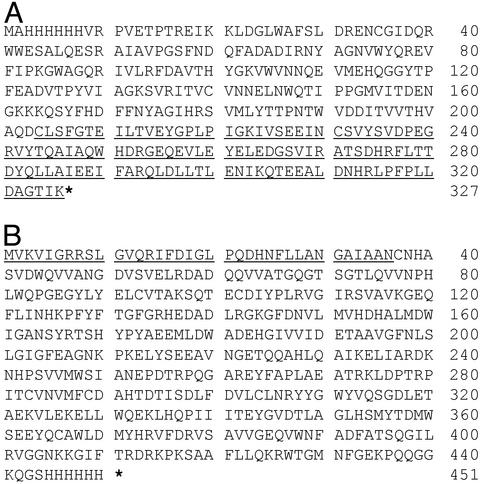
To monitor the protein trans-splicing reaction, the modified GUS gene that encodes a β-glucuronidase with both N- and C-terminal 6xHis tags was split and fused with Pint-n and Pint-c, respectively. The resulting sequences encoded for three fusion proteins. GUSn/Intn is an in-frame fusion between the first 203 amino acid residues of the tagged β-glucuronidase and the Int-n fragment of the DnaE intein (Fig. 1A). GUSn/Intn(6) has the same sequence as GUSn/Intn, except that the first six amino acid residues in the Int-n fragment were removed. Intc/GUSc is an in-frame fusion between the Int-c fragment of the DnaE intein and the C-terminal 415 amino acid residues of the tagged β-glucuronidase (Fig. 1B). The GUS gene split was designed so that the GUS-c fragment started with a Cys codon, which mimicked the natural split of the Ssp DnaE gene and was required for intein-mediated protein trans-splicing.
Figure 1.
Amino acid sequences of GUSn/Intn (A) and Intc/GUSc (B) fusion proteins. The underlined sequences indicate the Int-n fragment in GUSn/Intn fusion and the Int-c fragment in Intc/GUSc fusion. The stars represent stop codons. The 6xHis tags are located at N terminus of GUS-n fragment and C terminus of GUS-c fragment, respectively. Numbers of amino acid are indicated on the right. A deletion of the first six amino acid residues in the Int-n fragment of GUSn/Intn was introduced to yield GUSn/Intn(6).
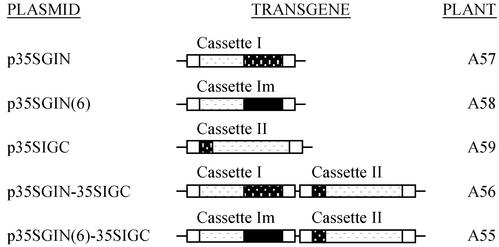
Three expression cassettes were constructed from the coding sequences of these in-frame fusions. They all had a cauliflower mosaic virus 35S promoter (35S-Pro) and a nopaline synthase terminator (NOS-Ter). Cassette I included a GUSn/Intn-coding sequence, cassette II included an Intc/GUSc-coding sequence, and cassette Im included a GUSn/Intn(6)-coding sequence, respectively. These cassettes were further integrated into the T-DNA regions of the binary vectors, either individually or in combination. This resulted in five expression plasmids: p35SGIN, p35SGIN(6), p35SIGC, p35SGIN-35SIGC, and p35SGIN(6)-35SIGC. The plasmids p35SGIN, p35SGIN(6), and p35SIGC contained single construct consisting of cassettee I, cassette Im, and cassette II, respectively. Whereas, p35SGIN-35SIGC contained a double construct consisting of cassette I plus cassette II and p35SGIN(6)-35SIGC contained cassette Im plus cassette II. The T-DNA region in p35SIGC contained an additional NOS-Pro∷Bar∷NOS-Ter cassette as a plant selection marker. The T-DNA regions in all other plasmids harbored NOS-Pro∷NPTII∷OCS-Ter to offer kanamycin resistance. The structures of these plasmids are summarized in Fig. 2.
Figure 2.
A summary of transgene constructs used for Arabidopsis transformation. Construct names, brief schemes, and titles of transfected plants are presented. Expression cassettes are named as indicated above each construct scheme. Cassette I, 35S-Pro∷GUSn/Intn∷NOS-Ter; cassette II, 35S-Pro∷Intc/GUSc∷NOS-Ter; cassette Im, 35S-Pro∷GUSn/Intn(6)∷NOS-Ter.
In Vivo Assembly of the Split β-Glucuronidase Through Protein Trans-Splicing.
To demonstrate catalytic activity of Ssp DnaE split intein and reconstitute a protein trans-splicing mechanism in plant cells, Agrobacteria carrying plasmid p35GIN-35SIGC was used to transform Arabidopsis. This plasmid has a T-DNA region containing expression cassettes I and II. It thus can integrate both cassettes into a single locus in Arabidopsis genomes to ensure coordinate production of GUSn/Intn and Intc/GUSc fusion proteins in the same cell. Plasmid p35GIN(6)-35SIGC was used as a control. This harbors expression cassettes Im and II to express GUSn/Intn(6) and Intc/GUSc fusion proteins. Because of a deletion of the first six amino acid residues in the Int-n part of the fusion, protein trans-splicing cannot be reconstituted.
Primary transgenic Arabidopsis plants were identified by germination of putative transformants on kanamycin selective medium. A55 and A56 represented transgenic plant lines, which had been transfected by constructs p35SGIN(6)-35SIGC and p35SGIN-35SIGC, respectively. The presence of the transgenes in the primary transgenic plants was confirmed by PCR assay (data not shown). Primary transformants were grown, and T2 seeds were collected from each plant.
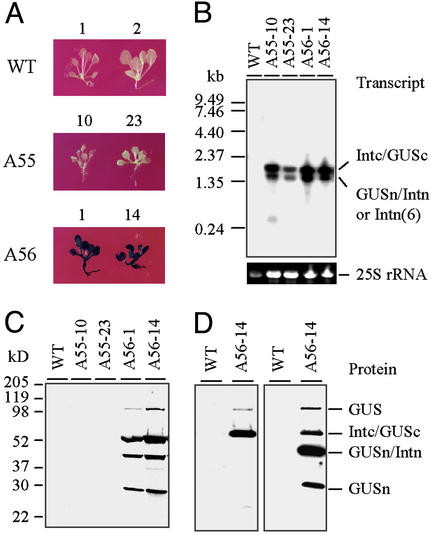
Protein trans-splicing was examined in T2 plants. For this, T2 seeds collected from A55 and A56 primary transgenic plants were germinated on kanamycin selective medium. WT Arabidopsis seeds were germinated on nonselective medium and used as background controls. Seedlings (2-wk-old) were analyzed by histochemical staining with X-Gluc to detect β-glucuronidase activity. The results obtained from two A55 events (A55-10 and A55-23) and two A56 events (A56-1 and A56-14) are shown in Fig. 3A to represent all examined A55 and A56 plants. The signature blue color of the cleaved X-Gluc appears throughout the A56-1 and A56-14 seedlings, implicating reassembly of β-glucuronidase from the split GUS-n and GUS-c protein fragments. A negative GUS staining result on the A55-10 and A55-23 seedlings indicates that intact β-glucuronidase cannot be assembled and that the partial β-glucuronidase encoded by GUS-n or GUS-c alone have no enzymatic activity.
Figure 3.
Examination of in vivo assembly of the divided β-glucuronidase fragments in 2-wk-old Arabidopsis seedlings. (A) GUS-staining assay. (B) RNA- filter hybridization assay. Total RNA (≈6 μg) was loaded in each lane and probed with 32P-labeled GUS-coding sequence. The 25S rRNA was used as a loading control in each lane. (C and D) Protein immunoblot assays. Total soluble protein (15 μg) was loaded in each lane. (C) Protein was probed with Penta-His Ab. (D) The protein was probed with anti-His(C Term)-HRP Ab at first (Left). After stripping, it was reprobed with Penta-His Ab (Right). For each assay, results obtained from two individual events of A55 and A56 transformations are shown. WT plants were used as controls.
The RNA expression profiles of A55, A56, and WT seedlings were inspected by RNA-filter hybridization assay. Transgene-specific RNAs were detected with a GUS gene probe. The results in Fig. 3B show that a 1.4-kb transcript from expression cassette Im and a 1.8-kb transcript from expression cassette II were detected in A55 RNA samples, and a 1.4-kb transcript from expression cassette I and a 1.8-kb transcript from expression cassette II were detected in A56 RNA samples. These results confirm normal transcription of the designed expression cassettes in both A55 and A56 plants. The fusion proteins and their splicing products in those seedlings were determined by immunoblot assay, using the Penta-His Ab. The results indicate that four proteins were detected from A56-1 and A56-14 seedlings (Fig. 3C). Based on their molecular weight and the fact that only GUS-n and GUS-c fragments possessed 6xHis tags, the top three proteins were assigned in order as the mature β-glucuronidase that was generated from the protein splicing, the Intc/GUSc fusion protein that was synthesized from cassette II, and the GUSn/Intn fusion protein that was synthesized from cassette I. These three proteins have calculated Mr values of 70.3, 50.7, and 37.4 kDa, respectively. To confirm this assignment, another immunoblot assay was performed on the protein extract of A56-14 seedlings. In this assay, the blot was probed with Anti-His(C Term)-HRP Ab at first and then reprobed with Penta-His Ab after stripping the first Ab away. Although the Penta-His Ab had revealed that four proteins contained one or more 6xHis tags (Fig. 3D Right), the Anti-His(C Term)-HRP Ab determined that only the top two proteins possessed a C-terminal 6xHis tag (Fig. 3D Left). Thus the two smaller proteins had only an N-terminal 6xHis tag. As a result, this assay further confirmed our assignment to the top three proteins. Based on its Mr and N-terminal His-tag, we proposed that the smallest protein was GUSn with a calculated Mr of 23.5 kDa, which may have been cleaved from GUSn/Intn fusion protein as a side reaction product of protein trans-splicing. It was surprising that the assay failed to detect His-tagged protein from protein extracts of A55 seedlings (Fig. 3C). In fact, neither mature β-glucuronidase nor GUSn/Intn(6) and Intc/GUSc fusion proteins accumulated in the seedlings although both expression cassette Im and II transcribed normally. This suggests that the GUSn/Intn(6) and Intc/GUSc fusion proteins may have been translated, but they are very unstable in plant cells.
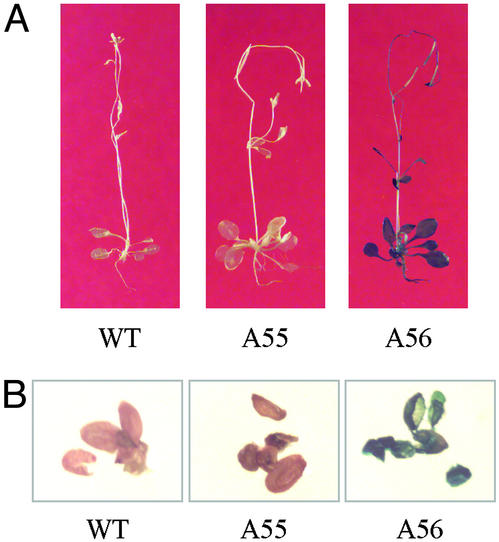
Intein-mediated protein trans-splicing requires optimal temperature, pH, and other conditions. Therefore, this process may not occur in all plant tissues. To further clarify this, the WT, A55, and A56 plants were grown in soil until they started producing seed, and then they were analyzed by a GUS histochemical-staining assay. The results show that β-glucuronidase activity is not present in any part of the WT and A55 plants but is in all parts of the A56 plant (Fig. 4A). Because seeds are protected within the seed pods, they are usually difficult to stain. To confirm the activity, young seeds were removed from the seed pods and stained. Fig. 4B shows that the result was consistent with that in whole plant. Thus we conclude that Ssp DnaE split intein can mediate protein trans-splicing in the most parts of a mature Arabidopsis, including root, stem, leaf, and seed. The splicing process not only ligates two halves of a split enzyme into an intact molecule but apparently also allows it to fold into its active form.
Figure 4.
Examination of in vivo assembly of the divided β-glucuronidase fragments in mature Arabidopsis plants. A55 and A56 Arabidopsis plants were subjected to a GUS-staining assay. WT Arabidopsis was used as control. (A) Entire Arabidopsis plants. (B) Young seeds.
Reconstitution of Protein Trans-Splicing Through Genetic Crossing.
These experiments demonstrate protein trans-splicing between two precursors that are synthesized from two different transgenes at the same locus in plant genomes. However, the splicing between transgenic products that are produced from different loci or chromosomes would have significant advantages for applications in transgenic plant platforms because the different transgenes could be brought together through classical plant breeding. To explore this hypothesis, A57, A58, and A59 transgenic plant lines were prepared by transfecting Arabidopsis with p35SGIN, p35SGIN(6), and p35SIGC, respectively. Homozygous plants of A57-6-3, A58-6-2, and A59-1-1 were identified after three rounds of selfing. For genetic crossing, the A59-1-1 plant was used as a pollen donor (♂), whereas A57-6-3 and A58-6-2 plants were used as pollen recipients (♀). The hybrids were confirmed by germination on the kanamycin-glufosinate selective medium and by PCR assays to reveal appropriate expression cassettes (data not shown). The hybrid plants were grown to maturity, and the second-generation self-fertilized seeds were collected from each of plants to represent individual crossing event for further analysis.
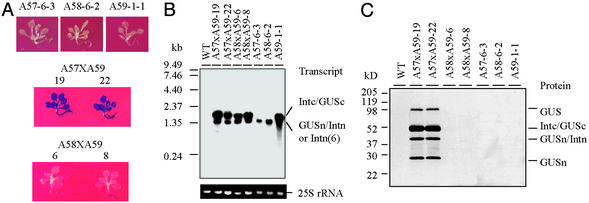
To inspect transgene expression in the hybrids, second-generation hybrid seeds of A57xA59 and A58xA59 were germinated on kanamycin-glufosinate selective medium. Meanwhile the homozygous seeds of A57-6-3 and A58-6-2 were germinated on kanamycin selective medium and A59-1-1 on glufosinate selective medium. WT seeds of Arabidopsis were germinated on nonselective medium, as background controls. Two-week-old seedlings were used in GUS histochemical staining, RNA preparation, RNA-filter hybridization, protein preparation, and immunoblot assay. The results obtained from two A57xA59 hybrids (A57xA59-19 and A57xA59-22) and two A58xA59 hybrids (A58xA59-6 and A58xA59-8) are summarized in Fig. 5 and are representative for each group of hybrids.
Figure 5.
Examination of protein trans-splicing in 2-wk-old Arabidopsis hybrids. (A) GUS-staining assay. (B) RNA-filter hybridization assay. Total RNA (≈6 μg) was loaded in each lane and probed with 32P-labeled GUS-coding sequence. The 25S rRNA was used as a loading control in each lane. (C) Protein immunoblot assay. Total soluble protein (15 μg) was loaded in each lane and probed with Penta-His Ab. In the assays, results obtained from two individual events of A57xA59 and A58xA59 crossings as well as that obtained from their homozygous parents are shown. WT plants were used as controls.
In the GUS histochemical-staining assays (Fig. 5A), the homozygous parents did not show any β-glucuronidase activity because they only harbor one of the expression cassettes. A57xA59 hybrids showed β-glucuronidase activity throughout the entire seedling but A58xA59 hybrids did not, indicating that protein trans-splicing between GUSn/Intn and Intc/GUSc fusion proteins was reconstituted and it required mediation of Ssp DnaE split inteins. It was interesting to note that β-glucuronidase activity was seen in the parts of roots of some A58xA59 hybrids. This implies that, under some special cellular conditions, the GUS fragments might functionally complement each other without splicing or the protein splicing might occur even when mutant intein elements were used. The conclusion drawn from the GUS histochemical staining was further confirmed by detailed molecular analysis. At first, RNA filter hybridization assays (Fig. 5B) demonstrated a normal RNA profile in each of the parent or hybrid plants. A57xA59 and A58xA59 hybrids synthesized two transcripts matching the RNA profile features of both their respective parents. In the immunoblot assays (Fig. 5C), A57xA59 hybrids showed accumulation of the fusion protein precursors and their splicing products, which included reassembled β-glucuronidase, Intc/GUSc precursor, GUSn/Intn precursor, and GUSn. But the A58xA59 hybrids didn't. This demonstrated that the intein-mediated protein trans-splicing was reconstituted in the A57xA59 hybrids. The fact that the fusion protein precursors did not accumulate in any of A57, A58, and A59 parental plants further confirmed our previous conclusion that those fusion proteins were translated but unstable in the plants where the protein splicing complexes could not form. In all assays, A58xA59 and A57xA59 hybrids showed results similar to those obtained from A55 and A56 plants, respectively. They indicate that chromosomal location of expression cassettes in the hybrids does not affect expression of the splicing components and demonstrate the feasibility of functional reconstitution of the protein trans-splicing mechanism through classic plant breeding.
Discussion
Although the mechanism of intein-mediated protein splicing has been studied since the early 1990s, it has yet to be demonstrated in higher plants. We reported here the in vivo reconstitution of a protein trans-splicing mechanism in Arabidopsis. Our results demonstrate that, in plants, the Ssp DnaE split intein is able to catalyze a molecular reassembly between two halves of β-glucuronidase. The reassembly results in a full-length β-glucuronidase with enzymatic activity, indicating accurate ligation and refolding of this enzyme.
To apply the protein trans-splicing in a plant platform, it is critical to ensure its efficiency and accuracy. Ssp DnaE intein was chosen in our experiments to ensure efficiency, because it is a naturally split intein and provides higher catalytic activity in moderate conditions than any known artificially split intein (12). The intein-coding sequences were also carefully designed and synthesized de novo, based on the rules of plant codon usage, to make them more compatible with plant gene expression machinery. To ensure accuracy of the reaction, the GUS gene was split between codons for Asp-203 and Cys-204 so that the GUS-c protein fragment started with a “native” Cys, rather than by the introduction of an extra Cys codon. GUS-n and GUS-c were directly fused to the N-end of Int-n and the C-end of Int-c, respectively, without any linker sequence between them. Therefore, an intact and native β-glucuronidase was recovered after protein splicing. In bacterial protein trans-splicing systems, short fragments of native Ssp DnaE extein were required as linkers between the Ssp DnaE intein and foreign extein to ensure the splicing reaction (11, 12). The plant protein splicing system did not require these linkers, indicating that Ssp DnaE intein may be more compatible with cellular conditions in plants. However, if the reassembled protein could tolerate integration of a few extra amino acids, the linker sequences may further increase the efficiency of protein trans-splicing in plant system.
It is still unknown how to directly control and regulate the protein splicing. Our data has shown that Ssp DnaE split intein actively catalyzes protein splicing reactions throughout the entire plant, including many agriculturally important tissues. Thus, it is possible to construct intein–extein fusion under control of tissue-specific, development-specific, or chemical inducible promoters to achieve conditional reconstitution of protein trans-splicing in plant cells. We also have demonstrated that different components of the protein trans-splicing machinery can be stacked in the parental plants separately. The splicing can be reconstituted in hybrid progenies by bringing the components together through genetic crossing. In this approach, the protein trans-splicing can be restricted in a specific generation.
It was surprising to learn that partial β-glucuronidase, when fused with intein, was apparently unstable in plant cells. It is unclear whether truncation of β-glucuronidase or integration of split intein caused the instability. The results from A56 and A57xA59 plants showed that the GUSn/Intn and Intc/GUSc fusion proteins could survive in the plants where the protein trans-splicing was successfully reconstituted. A strong affinity between Ssp DnaE Int-n and Int-c has been reported in previous work (26). We propose that interaction between the two halves of the intein may result in a splicing complex, which not only ensures a protein splicing reaction but also protects the protein precursors from degradation. It was also unexpected that β-glucuronidase activity was detected in the parts of roots in several A58xA59 seedlings. In these seedlings, a 6-aa fragment including a splicing required Cys was missing from Int-n part of GUSn/Intn fusion protein. The mutated Int-n could not catalyze protein splicing, but it might still have some affinity to weakly associate with Intc/GUSc, thus resulting in a complementation of β-glucuronidase when the cellular conditions allowed. On the other hand, an alternative protein splicing mechanism has been identified for KlbA intein, which no longer required this Cys (27). It is also possible that a similar alternative mechanism was operating in these root cells.
Because of its capability of seamlessly ligating two separate protein fragments together without leaving any footprint, protein trans-splicing has great potential in plant genetic engineering. One area of application is to use it as a “molecular switch” to turn on a gene expression mechanism or a metabolic pathway through reassembly of gene regulators or metabolic enzymes. Combined with plant breeding techniques or specific promoters, the mechanism and pathway can be activated in a designated generation, tissue, developmental stage, or with special stimulators. Protein trans-splicing may limit the environmental impact of herbicide resistance genes by keeping different parts of the gene in plastid and nuclear genomes while assembling gene products in cytosol (11). Another attractive area is to use inteins to produce larger and more complex proteins. For example, the larger a heterologous protein or enzyme is usually more difficult, and sometimes impossible, to synthesize in plants or other organisms. If protein trans-splicing can be integrated into a production platform, however, smaller fragments of the protein could be synthesized and then assembled into a complete molecule. Advanced protein polymer synthesis often requires assembly of the building blocks, which include different functional domains and peptide backbones (28). By using the protein trans-splicing mechanism, these building blocks could be stacked in separate transgenic traits, and assembled into the protein polymer in the hybrid progenies through genetic crossing.
Acknowledgments
We thank Dr. Patrick Ireland and Dr. Timothy Caspar for advice and Dr. Arthur Hunt for comments on the manuscript. This work was supported by DuPont Central Research and Development as part of the research program in the plant gene expression group.
Abbreviations
- Ssp
Synechocystis sp. PCC6803
- DnaE
catalytic subunit α of DNA polymerase III
- Int-n
N-terminal half of split intein
- Int-c
C-terminal half of split intein
- NOS-Ter
nopaline synthase terminator
- 35S-Pro
35S promoter
- HRP
horseradish peroxidase
- GUS
β-glucuronidase
Footnotes
References
- 1.Perler F B. Cell. 1998;92:1–4. doi: 10.1016/s0092-8674(00)80892-2. [DOI] [PubMed] [Google Scholar]
- 2.Noren C J, Wang J, Perler F B. Angew Chem Int Ed. 2000;39:450–466. [PubMed] [Google Scholar]
- 3.Chong S, Xu M-Q. J Biol Chem. 1997;272:15587–15590. doi: 10.1074/jbc.272.25.15587. [DOI] [PubMed] [Google Scholar]
- 4.Shingledecker K, Jiang S, Paulus H. Gene. 1998;207:187–195. doi: 10.1016/s0378-1119(97)00624-0. [DOI] [PubMed] [Google Scholar]
- 5.Perler F B. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills K V, Lew B M, Jiang S-Q, Paulus H. Proc Natl Acad Sci USA. 1998;95:3543–3548. doi: 10.1073/pnas.95.7.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Southworth M W, Adam E, Panne D, Byer R, Kautz R, Perler F B. EMBO J. 1998;17:918–926. doi: 10.1093/emboj/17.4.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, Xu M-Q, Liu X-Q. Biochim Biophys Acta. 1998;1387:422–432. doi: 10.1016/s0167-4838(98)00157-5. [DOI] [PubMed] [Google Scholar]
- 9.Yamazaki T, Otomo T, Oda N, Kyogoku Y, Uegaki K, Ito N, Ishino Y, Nakamura H. J Am Chem Soc. 1998;120:5591–5592. [Google Scholar]
- 10.Wu H, Hu Z, Liu X-Q. Proc Natl Acad Sci USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Ghosh I, Paulus H, Xu M-Q. Appl Environ Microbiol. 2001;67:1025–1029. doi: 10.1128/AEM.67.3.1025-1029.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin D D, Xu M-Q, Evans T C., Jr Biochemistry. 2001;40:1393–1402. doi: 10.1021/bi001786g. [DOI] [PubMed] [Google Scholar]
- 13.Scott C P, Abel-Santos E, Wall M, Wahnon D C, Benkovic S J. Proc Natl Acad Sci USA. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y. Anal Chem. 2001;73:2516–2521. doi: 10.1021/ac0013296. [DOI] [PubMed] [Google Scholar]
- 15.Kinsella T M, Ohashi C T, Harder A G, Yam G C, Li W, Peelle B, Pali E S, Bennett M K, Molineaux S M, Anderson D A, et al. J Biol Chem. 2002;277:37512–37518. doi: 10.1074/jbc.M206162200. [DOI] [PubMed] [Google Scholar]
- 16.Prado F, Aguilera A. Curr Genet. 1994;25:180–183. doi: 10.1007/BF00309546. [DOI] [PubMed] [Google Scholar]
- 17.Oldenburg K R, Vo K T, Michaelis S, Paddon C. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jefferson R A, Burgess S M, Hirsh D. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu, Z.-B. & Odell, J. T. (1999) U.S. Patent 5,968,793.
- 20.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 21.Crow J F. Genetics Notes. 7th Ed. Minneapolis: Burgess; 1976. pp. 229–235. [Google Scholar]
- 22.Jefferson R A. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 23.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Schuster G, Stern D B. Plant Cell. 1996;8:1409–1420. doi: 10.1105/tpc.8.8.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray E E, Lotzer J, Eberle M. Nucleic Acids Res. 1989;17:477–498. doi: 10.1093/nar/17.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Pradhan S, Evans T C., Jr Gene. 2001;263:39–48. doi: 10.1016/s0378-1119(00)00568-0. [DOI] [PubMed] [Google Scholar]
- 27.Southworth M W, Benner J, Perler F B. EMBO J. 2000;19:5019–5026. doi: 10.1093/emboj/19.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Brien J P, Fahnestock S R, Termonia Y, Gardner K H. Adv Mater. 1998;10:1185–1195. [Google Scholar]