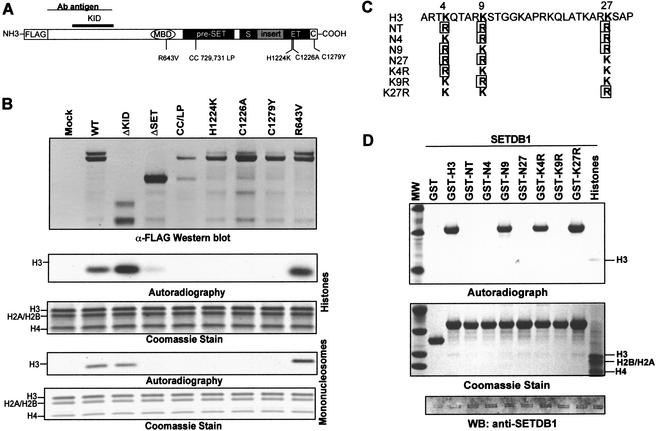
Figure 3.
SETDB1 selectively methylates Lys 9 of histone H3. (A) Schematic illustrating the relative position of single amino acid substitutions synthetically introduced into the MBD, pre-SET, SET, and post-SET domains as indicated. (B) Deletion of the post-SET and part of the SET homologies and single amino acid mutations at highly conserved residues within the catalytic domain (pre-SET, SET, and post-SET) impair the H3-methylase activity of SETDB1. The anti-Flag Western blot confirms the expression and Flag immunopurification of the indicated proteins. The ΔKID and ΔSET proteins correspond to amino acids 570–1291 and 1–951 of SETDB1, respectively. (C) Amino acid sequence of the N-terminal tail of histone H3 (1–30) is shown at the top with the K4, K9, and K27 residues highlighted. The various lysine to arginine mutations in K4, K9, and K27 derived to determine the substrate specificity of SETDB1 are indicated. All lysine (K) to arginine (R) mutations at K4, K9, and K27 are boxed. (D) The SETDB1 methyltransferase activity is highly specific for K9. Five micrograms of the corresponding GST–H3N protein was used as substrate in the in vitro HMTase assay with Flag-purified SETDB1 (Fig. 1D). Coomassie blue stain shows the purified GST–histone H3 substrates. Autoradiograph shows corresponding [3H]methyl-labeled products. Western blot confirms presence of Flag-SETDB1 in the HMTase reaction.