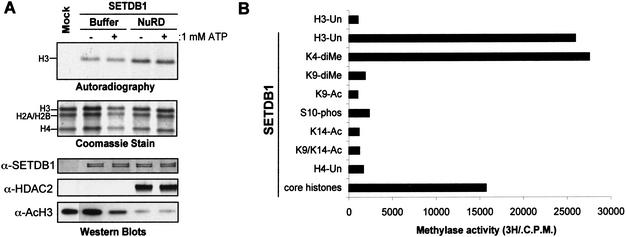
Figure 4.
Dissecting the histone code and methylation by SETDB1. (A) Pretreatment of a core histone substrate with a homogenously pure histone deacetylase complex, NuRD, enhanced methylation of histone H3 in vitro, concomitantly with deacetylation of histone H3 (anti-AcH3 Western blot). Coomassie blue stain shows equal loading of histone proteins. Autoradiograph shows corresponding [3H]methyl-labeled products. Anti-SETDB1 and anti-HDAC2 Western blots show the presence of SETDB1 and HDACs in the corresponding HMTase reactions. (B) Effect of histone modifications on the enzymatic activity of SETDB1. One microgram of unmodified or acetylated (K9-Ac, K14-Ac, K9,K14-Ac), phosphorylated (S10-phos), or methylated (K4-diMe, K9-diMe) peptides corresponding to the N-terminal tail of histone H3 and H4 were used as substrates in the in vitro methylation assay with Flag-purified SETDB1. Methylation was quantified via a filter binding assay and represented as raw counts per minute (C.P.M.) incorporated.