Abstract
We detected, for the first time, the occurrence of vegetative incompatibility between different isolates of the arbuscular mycorrhizal fungal species Glomus mosseae. Vegetative compatibility tests performed on germlings belonging to the same isolate showed that six geographically different isolates were capable of self-anastomosing, and that the percentage of hyphal contacts leading to fusions ranged from 60 to 85%. Successful anastomoses were characterized by complete fusion of hyphal walls, protoplasm continuity and occurrence of nuclei in the middle of hyphal bridges. No anastomoses could be detected between hyphae belonging to different isolates, which intersected without any reaction in 49 to 68% of contacts. Microscopic examinations detected hyphal incompatibility responses in diverse pairings, consisting of protoplasm retraction from the tips and septum formation in the approaching hyphae, even before physical contact with neighboring hyphae. Interestingly, many hyphal tips showed precontact tropism, suggesting that specific recognition signals may be involved during this stage. The intraspecific genetic diversity of G. mosseae revealed by vegetative compatibility tests was confirmed by total protein profiles and internal transcribed spacer-restriction fragment length polymorphism profiles, which evidenced a higher level of molecular diversity between the two European isolates IMA1 and BEG25 than between IMA1 and the two American isolates. Since arbuscular mycorrhizal fungi lack a tractable genetic system, vegetative compatibility tests may represent an easy assay for the detection of genetically different mycelia and an additional powerful tool for investigating the population structure and genetics of these obligate symbionts.
Arbuscular mycorrhizal (AM) fungi are key microorganisms of the soil-plant system that are fundamental for soil fertility and plant nutrition (47). They live in symbiosis with the roots of most plant species and produce spores in the soil, which are able to germinate in the absence of host-derived signals but unable to complete their life cycle without establishing a functional symbiosis with a host plant (16). Under experimental conditions, hyphae developing from germinated spores in the absence of the host elongate and give rise to coenocytic mycelia whose maximum extension ranges from 71 mm (Glomus species) to 544 mm (Gigaspora species) (12, 31). When hyphae of the same isolate come into contact, they form anastomoses, which represent the fundamental mechanism allowing the development of large mycelial networks in presymbiotic and symbiotic mycelia (15, 18). Anastomoses have been shown to occur widely between hyphae belonging to the same and different germlings of the same isolates in Glomus mosseae, Glomus caledonium, Glomus intraradices, whereas hyphae belonging to different species never fused, suggesting fungal ability to recognize species level differences (15, 19, 49). The high frequency of anastomosis formation and the establishment of protoplasmic continuity between fused hyphae suggested that the ability of self-compatible hyphae to recognize each other could represent a means for the exchange of genetic material, particularly if anastomoses occurred between hyphae derived from genetically different spores (15, 26). Several studies have reported that individual spores of AM fungi, which are multinucleate (they may contain thousands of nuclei [7, 53]) show a high level of genetic diversity in the internal transcribed spacer (ITS) region of the nuclear rRNA genes (3, 28, 30, 42). Since AM fungi are considered clonal organisms (40), anastomoses between different germlings may represent a mechanism for the maintenance of genetic diversity in the absence of sexual recombination (6, 41). However, so far nothing is known of hyphal compatibility between conspecific isolates, which could allow genetic exchange through anastomosis formation.
Vegetative compatibility tests have been utilized to investigate the occurrence of different isolates within defined geographical areas and to study the population structure of different pathogenic, saprophytic, and ectomycorrhizal fungi. Such tests, performed by scoring hyphal interactions directly, revealed the occurrence of isolates belonging to different vegetative compatibility groups (VCGs) within the same fungal species (9, 10, 13, 20, 21, 29, 33, 44, 45, 54). Moreover, different experimental works reported that vegetative compatibility tests were often consistent with genetic and biochemical analyses and that genetic diversity was generally greater between isolates belonging to different VCGs (8, 32, 44, 48).
In this work we investigated the genetic relationships among isolates of the AM fungal species Glomus mosseae originating from different geographical regions in Europe and the United States. The experiments were aimed at (i) detecting the occurrence of intraspecific vegetative compatibility, (ii) determining the biochemical diversity of isolates by comparing isozyme and total protein profiles, and (iii) determining intraspecific molecular diversity by comparing restriction fragment length polymorphisms (RFLP) of PCR-amplified ITS regions of the ribosomal DNA (rDNA).
MATERIALS AND METHODS
Fungal material.
The AM fungi used were geographically different isolates belonging to the species G. mosseae (Nicolson et Gerdemann) Gerdemann et Trappe. Details of the isolates are given in Table 1. The AM fungal species G. caledonium (Nicolson et Gerdemann) Trappe et Gerdemann (Rothamsted isolate = BEG20 = IMA2) and Glomus coronatum Giovannetti (BEG28 = IMA3) were used for comparison in some experiments. Spores of G. mosseae isolates AZ225C, BEG25, IMA1, IN101C, and SY710 and of G. caledonium and G. coronatum were obtained from pot cultures maintained in the collection of the Department of Chemistry and Agricultural Biotechnology, University of Pisa, Pisa, Italy. Spores of G. mosseae isolate BEG69 were kindly provided by Corinne Leyval, INRA, Nancy, France.
TABLE 1.
Geographic origin and inoculum source of AM fungal isolates used in the present work
| Fungal species | Isolate code | Geographic origin | Collector | Original inoculum supplierc |
|---|---|---|---|---|
| G. mosseae | AZ225C | Arizona | J. C. Stutz | INVAM, Morgantown, W.Va. |
| G. mosseae | BEG25a | West Sussex, United Kingdom | J. C. Dodd | IIB, Canterbury, United Kingdom |
| G. mosseae | BEG69 | Pas-de-Calais, France | C. Leyval | CPB, Nancy, France |
| G. mosseae | IMA1b | Kent, United Kingdom | B. Mosse | Rothamsted Exp. St., United Kingdom |
| G. mosseae | IN101C | Indiana | R. Kemery | INVAM, Morgantown, W.Va. |
| G. mosseae | SY710 | Syria | D. Sands | INVAM, Morgantown, W.Va. |
| G. caledonium | IMA2 | Hartfordshire, United Kingdom | D. Hayman | Rothamsted Exp. St., United Kingdom |
| G. coronatum | IMA3 | Tuscany, Italy | M. Giovannetti | IMA, Pisa, Italy |
BEG, Bank of European Glomales.
IMA, International Microbial Archives.
Abbreviations: INVAM, International Culture Collection of Arbuscular and Vesicular-Arbuscular Mycorrhizal Fungi; IIB, International Institute of Biotechnology; CPB, Centre de Pedologie Biologique (CNRS); Exp. St., Experimental Station.
Spore collection.
Spores were extracted from pot culture soil by wet sieving and decanting, down to a mesh size of 100 μm. Spores retained on sieves were flushed into petri dishes and manually collected with a capillary pipette under a dissecting microscope. Only intact, healthy spores were selected. For biochemical and molecular analyses, spores were placed in an Eppendorf tube after two sonication steps (60 s each) in a B-1210 cleaner (Branson Ultrasonics, Soest, The Netherlands) and washed three times in sterile distilled water.
Morphological characterization.
All G. mosseae isolates were morphologically identified by using species diagnostic characteristics described by Gerdemann and Trappe (14). At least 50 individual spores were mounted on microscope slides and examined under a Polyvar light microscope equipped with Nomarski differential interference contact optics (Reichert-Young, Vienna, Austria). The spores were observed and measured by using Quantimet 500 image analysis software (Leica, Milan, Italy).
In vitro spore germination.
The different G. mosseae isolates were tested for their ability to germinate and grow in vitro. The following G. mosseae isolates were used: AZ225C, BEG25, BEG69, IMA1, IN101C, and SY710. Spores were surface sterilized with 2% chloramine T supplemented with streptomycin (400 μg/ml) for 20 min, rinsed five times in sterile distilled water (SDW), and placed on sterile cellophane membranes (20 by 20 mm; Hoefer, San Francisco, Calif.) laid on 1% water agar in 9-cm-diameter petri dishes. At least 10 replicate membranes were prepared for each isolate. Plates were sealed with Parafilm and incubated at 28°C in the dark. After 14 to 32 days' incubation, spore germination was assessed under a Wild dissecting microscope (Leica).
In vivo spore germination.
The different G. mosseae isolates were tested for their ability to germinate and grow in vivo. Spores or sporocarps were washed by vortexing in SDW for 20 s, rinsed three times in SDW, and then placed on 47-mm-diameter cellulose nitrate Millipore membranes (pore diameter, 0.45 μm). Membranes were placed on a moistened membrane of the same type laid on sterile quartz grit in 14-cm-diameter petri dishes, which were sealed with Parafilm and incubated at 28°C in the dark. After 21 days' incubation, spore germination was assessed under a Wild dissecting microscope after staining with 0.05% trypan blue in lactic acid. Membranes were then mounted on microscope slides for growth measurements, which were carried out under the Polyvar microscope, by using Quantimet 500 image analysis software.
Occurrence and frequency of anastomoses.
The in vivo experimental system and the following G. mosseae isolates were used: AZ225C, BEG25, BEG69, IMA1, IN101C, SY710. Spores or sporocarps were washed and placed on Millipore membranes as described above. Two spores or sporocarps belonging either to the same or to different isolates were paired on each membrane approximately 1 cm apart and at least 30 replicate membranes were prepared for each pairing. Membranes were placed on a moistened membrane of the same type laid on sterile quartz grit in 14-cm-diameter petri dishes, which were sealed with Parafilm and incubated at 28°C in the dark. After 21 to 42 days' incubation, occurrence of anastomosis was assessed on germlings by staining for the presence of succinate dehydrogenase (SDH) activity (15, 46). Deposition of formazan salts in hyphae allowed the visualization of viable mycelia and of protoplasmic continuity between fusing hyphae. Membranes bearing stained germlings were mounted on microscope slides using 0.05% trypan blue in lactic acid and observed under the Polyvar microscope. All hyphal fusions were counted at magnifications of ×125 to 500 and verified at a magnification of ×1,250. Frequency of anastomosis was calculated by determining the proportion of hyphal contacts that led to hyphal fusions. Three independent replicate experiments were performed.
To visualize the occurrence and location of nuclei in anastomosed hyphae, some membranes were stained with diamidinophenylindole (DAPI) (5 μg/ml) in a 1:1 (vol/vol) water-glycerol solution and observed under epifluorescence with the Polyvar microscope using the filter combination U1 (15).
To visualize hyphal interactions, some membranes were stained with DAPI and mounted in a 0.01% (wt/vol) solution of Calcofluor White (Sigma Aldrich s.r.l., Milan, Italy) as described by Sbrana et al. (43) and observed under epifluorescence with the filter combination U1.
Isozyme analysis.
The G. mosseae isolates AZ225C, BEG25, IMA1, IN101C, and SY710 and the isolate IMA2 were used. Soluble protein extracts from about 15 to 30 spores were transferred to Eppendorf tubes and processed as previously described (39). Briefly, spores were crushed in sucrose-Triton extraction buffer (1.5 spores/μl) and centrifuged for 20 min at 4°C (12,500 × g). The supernatants were immediately loaded into wells and run on vertical nondenaturing polyacrylamide gels (4% [wt/vol] stacking gel and 7.5% [wt/vol] separating gel) at 4°C in a Hoefer mini-gel system in an electrode buffer containing 25 mM Tris HCl (pH 8.3) and 192 mM glycine, at a constant current of 15 mA per gel. Isozymes for malate dehydrogenase (MDH) (EC 1.1.1.37) and esterase (EST) (EC 3.1.1.1) were visualized after staining as described by Rosendahl and Sen (39). Gel images were stored as TIFF files by using the ImageMaster VDS system (Amersham Biosciences, Europe GmbH).
SDS-PAGE analysis.
The G. mosseae isolates AZ225C, BEG25, IMA1, and IN101C and the G. coronatum isolate IMA3 were used. The number of spores utilized for each isolate ranged between 60 (for AZ225C) and 80 (for IMA1), in order to yield the same quantity of protein extracts. Protein extraction was carried out as previously described (4). Spores were ground at 4°C, and soluble proteins were extracted in a buffer containing 62.5 mM Tris-HCl (pH 6.8), 2 mM phenylmethylsulfonyl fluoride, 290 mM mercaptoethanol, 20% glycerol, and 2% sodium dodecyl sulfate (SDS). Samples were boiled at 100°C for 10 min, centrifuged at 12,500 × g for 20 min at 4°C, and loaded immediately onto gels, adding 0.002% bromophenol blue. One-dimensional SDS-polyacrylamide gel electrophoresis (PAGE) was carried out using a discontinuous buffer system (27). Extracts were loaded onto constant gradient gels (12%) and run in a Mini Protean II slab cell system (Bio-Rad Laboratories, s.r.l., Milan, Italy) at 25 mA and 200 V at room temperature. Gels were first treated with Brilliant Blue R-250 (Sigma-Aldrich s.r.l.) and then stained with a silver staining kit from Bio-Rad, photographed, and air dried in cellulose sheets.
Gel images were stored as TIFF files by using the ImageMaster VDS system and analyzed with ImageMaster 1D Elite software (Amersham Biosciences, Europe GmbH). Only bands between 66 and 14.5 kDa were analyzed. The dendrogram was generated by the software after matching lanes using the Dice similarity index and the unweighted pair-group arithmetic average (UPGMA) clustering method. Both calculations of cophenetic correlation coefficient and bootstrapping analysis using 100 replicate data sets were performed, to assess the significance of the groupings obtained by cluster analysis (38, 52).
DNA amplification and restriction enzyme analysis.
The G. mosseae isolates AZ225C, BEG25, IMA1, and IN101C and the isolate IMA3 were used. DNA extracts were prepared according to the protocol described by Redecker et al. (37). Briefly, a batch of spores (10 to 20) was crushed in Eppendorf tubes using a glass pestle in 4 μl of 0.25 M NaOH and incubated in boiling water (1 min). After adding 2 μl of 0.5 M Tris-HCl (pH 8.0) and 4 μl of 0.25 M HCl, the extract was dipped again in boiling water (2 min). The PCR amplification was carried out in a 50-μl final volume containing 2 μl of DNA extract. Final concentrations of PCR mix components were 0.1 mM (each) dATP, dCTP, dGTP, and dTTP (Finnzymes, Celbio s.r.l., Milan, Italy); 0.2 μM (each) ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) primers; 50 mM KCl; 10 mM Tris-HCl (pH 8.3); 0.1% Triton X-100; 4.0 mM MgCl2; and 0.025 U of Taq DNA polymerase per μl (Finnzymes). A thermal cycler (GeneAmp PCR system 2400; Applied Biosystems, Milan, Italy) was programmed as described by Redecker et al. (37).
Amplified DNA was purified using the Amicon Microcon-PCR device (Millipore), and aliquots of 2 μl were digested separately with restriction enzymes AluI, DpnII, EcoRI, HaeIII, HinfI, MspI, RsaI, and TaqI according to the manufacturer's instructions (BioLabs, Hitchin, United Kingdom). Digested DNA was run through 4% agarose containing ethidium bromide (0.5 ppm). For electrophoresis, 20 μl of digested sample was mixed with 3 μl of tracking buffer (30% glycerol, 0.125% bromophenol blue, and 20 mM Tris-HCl, pH 8.0) and loaded onto a gel with 1× Tris-borate-EDTA as the running buffer. Electrophoresis was carried out at 100 V on a submerged horizontal gel electrophoresis system (Bio-Rad). A DNA molecular weight marker (100-bp ladder; BioLabs) was used as a sizing marker. DNA profiles were acquired with the ImageMaster VDS system and analyzed using ImageMaster Elite software.
The presence or absence of bands for each restriction enzyme was coded in binary form (as 0 or 1). The data were then combined in a pooled matrix used to perform cluster analysis applying the Dice similarity index and the UPGMA algorithm. Both calculations of cophenetic correlation coefficient and bootstrapping analysis using 100 replicate data sets were performed to assess the significance of the groupings obtained by cluster analysis (38, 52). A dendrogram was generated by TREECON for Windows software (52).
RESULTS
Morphological characterization.
Morphological characterization of spores and sporocarps of G. mosseae isolates AZ225C, BEG25, BEG69, IMA1, IN101C, and SY710 showed that their features were consistent with the original description of the species (14). Sporocarps were not produced by isolates IN101C and BEG69, which produced only ectocarpic spores. The number of spores per sporocarp ranged from 1 to 4 in AZ225C, BEG25, and SY710 isolates and from 10 to 12 in IMA1. Spore diameters ranged from 152 ± 5.8 μm (mean ± standard error of the mean) in BEG69 to 256 ± 3.5 μm in IN101C.
Spore germination and mycelial growth.
Spores and/or sporocarps of the six G. mosseae isolates showed different germination abilities, depending on the experimental system. Surface-sterilized spores grown in vitro exhibited the lowest germination percentages in all isolates, and some of these (BEG25, IN101C, and SY710) did not germinate at all in axenic culture. In contrast, spore germination was higher in all isolates when the in vivo culture system was used (Table 2). From some sporocarps of isolate SY710 thin, pigmented and septate hyphae developed, showing fungal contamination.
TABLE 2.
Spore and/or sporocarp germination percentages of different isolates belonging to the AM fungal species G. mosseae
| Isolate | % Germination (95% CIa)
|
||
|---|---|---|---|
| In vitro spores | In vivo spores | In vivo sporocarps | |
| AZ225C | 28 (12.1-49.4) | 32 (10.5-27.3) | 49 (37-60) |
| BEG25 | 0 (0-13.7) | 32 (22.7-42.9) | 50 (33.8-66.2) |
| BEG69 | 24 (9.4-45.1) | 28 (16.2-42.5) | |
| IMA1 | 12 (2.5-31.2) | 17 (9.6-26) | 43 (27-59.1) |
| IN101C | 0 (0-13.7) | 30 (20.8-40.6) | |
| SY710 | 0 (0-13.7) | 8 (3.2-15.4) | 5 (0.6-16.9) |
Numbers in parentheses indicate 95% confidence intervals (CI).
Hyphal growth per germinated spore, assessed in vivo, varied with the different isolates and ranged from 34.5 ± 3.5 mm (mean ± standard error of the mean) and 35.9 ± 2.9 in BEG69 and SY710, respectively, to 119.5 ± 14.4 mm in IMA1.
Given the low germinability of some isolates in vitro, anastomosis data were collected only from in vivo experiments.
Occurrence and frequency of anastomoses within the same isolate.
Anastomosis formation between different spores of the same isolate was detected in all G. mosseae isolates. Hyphae originating from germinated spores or sporocarps paired on membranes came into contact after 21 to 42 days' incubation, depending on individual growth rates. Successful anastomoses were characterized by complete fusion of hyphal walls. Protoplasm continuity, the characteristic feature of true anastomosis, evidenced by formazan salt depositions in hyphal bridges (SDH activity), was consistently achieved in all the anastomoses observed.
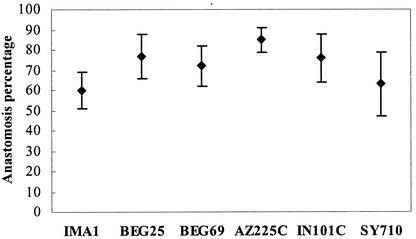
DAPI staining and fluorescence microscopy detected many nuclei in the middle of hyphal bridges formed between anastomosing hyphae. No hyphal incompatibility reactions such as vacuolization, septum development, or hyphal lysis were detected after hyphal fusions. High percentages of anastomoses occurred in all isolates, ranging from 60% in IMA1 (55 fusions, 91 contacts) to 85% in AZ225C (206 fusions, 242 contacts) (Fig. 1). During hyphal interactions between spores belonging to the same isolate which did not lead to anastomosis, hyphae showed no contact interference.
FIG. 1.
Percentages of successful anastomoses with respect to total hyphal contacts in the different Glomus mosseae isolates. Bars represent 95% confidence limits.
Interactions between different isolates.
No anastomoses were detected in pairings between germlings of different isolates of G. mosseae (0 fusions, 473 hyphal contacts). In particular, 95% confidence limits were calculated for each pairing and were 0 to 3 for AZ225C-IN101C, 0 to 14 for AZ225C-BEG25, 0 to 9 for AZ225C-BEG69, 0 to 12 for AZ225C-IMA1, 0 to 3 for IMA1-BEG25, 0 to 13 for IMA1-IN101C, 0 to 12 for IN101C-SY710, 0 to 23 for IN101C-BEG25, 0 to 23 for AZ225C-SY710, and 0 to 19 for IMA1-SY710. Due to poor mycelial growth of SY710 and BEG69 isolates, the pairings BEG69-IMA1, BEG69-BEG25, BEG69-IN101C, BEG69-SY710, and SY710-BEG25 produced a low number of hyphal contacts and confidence limits are not shown.
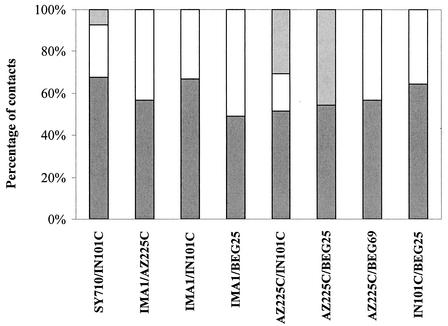
During interisolate hyphal interactions, hyphae appeared to intersect without any reaction in 49 to 68% of contacts (Fig. 2). Incompatibility reactions were detected in the following pairings: IMA1-BEG25 (in 51% of contacts), AZ225C-IN101C (49%), AZ225C-BEG25 (46%), IMA1-AZ225C and AZ225C-BEG69 (43%), IN101C-BEG25 (36%), IMA1-IN101C (33%), and SY710-IN101C (32%). The main feature of incompatibility responses was represented by protoplasm retraction and septum formation in the approaching hyphae, prior to or after physical contact with neighboring hyphae (Fig. 3a and b). Interestingly, some hyphal tips showed precontact tropism and growth reorientation (Fig. 3c). In many contacts hyphal swellings produced by the approaching hypha on the surface of the contacted one were characterized by localized wall thickenings, which were detected by Calcofluor staining (Fig. 4a). After this early incompatible response, protoplasm was withdrawn and consecutive septa were formed. No wall lysis was observed and the apical traits of contacting hyphae appeared empty and septate (Fig. 3d).
FIG. 2.
Percentage of hyphal contacts leading to hyphal intersection with no cellular response (dark gray bars), type A interaction between hyphae (open bars), and type B interaction between hyphae (light gray bars). See text for description of interactions of types A and B.
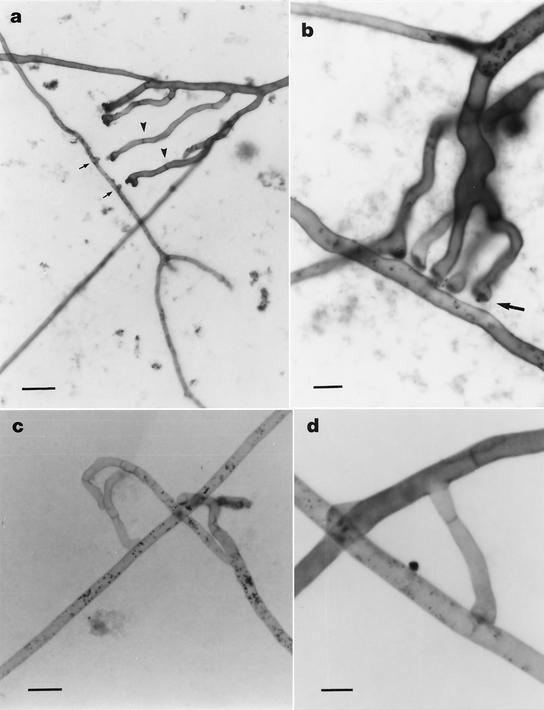
FIG. 3.
Light micrographs showing incompatible interactions between hyphae belonging to geographically different isolates of the AM fungus G. mosseae, after SDH and trypan blue staining. (a) Development of multiple hyphal branches (IN101C) growing towards branch initiation sites (arrows) on the recipient hypha (AZ225C). Note hyphal swellings and consecutive retraction septa (arrowheads) produced prior to any physical contact between the hyphae. Scale bar = 35 μm. (b) Multiple hyphal branches (AZ225C) showing precontact protoplasm retraction and localized thickening of hyphal tips (arrow). Scale bar = 9 μm. (c) Change of growth direction in an approaching hypha (BEG25) accompanied by protoplasm withdrawal and septum formation. Scale bar = 17 μm. (d) Retraction septa developed by an approaching hypha (IN101C) after incompatible interaction (type A). Scale bar = 9 μm.
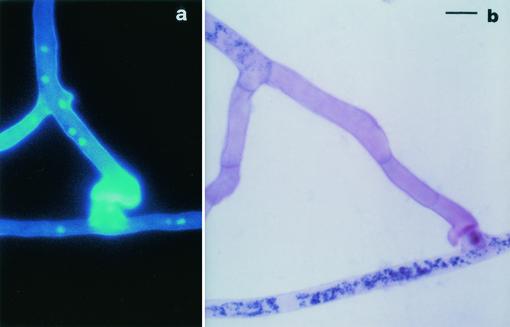
FIG. 4.
Type B incompatible interactions between geographically different isolates of the AM fungus G. mosseae. Scale bar = 9 μm. (a) Epifluorescence image, after Calcofluor and DAPI staining, showing wall thickening of a hyphal swelling developed by the approaching hypha (AZ225C) on a lateral branch initial of the contacted one (IN101C). (b) Light micrograph, after SDH and trypan blue staining, showing protoplasm withdrawal and septum formation in an approaching hypha (IN101C) after incompatible contact with a branch initial (SY710).
Two different types of incompatible interactions were detected: in type A an apical swelling was differentiated by the approaching hypha on the contacted one (Fig. 3c and d); in type B the approaching hyphal tip developed single or multiple branches growing towards the recipient hypha, which produced one or many branch initiation sites on lateral wall (Fig. 3a and 4a and b). Interactions of type A were the most frequent in all pairings analyzed, with the exception of AZ225C-IN101C and AZ225C-BEG25 pairings, where responses of type B occurred in 63 and 100% of contacts involving incompatible interactions (Fig. 2).
Isozyme and protein analysis.
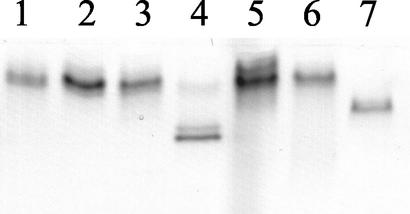
The patterns of MDH were well resolved and reproducible: all G. mosseae isolates showed a single band with no variation in intensity and position, except for isolate SY710, which showed two additional bands, confirming contamination observed during germination tests. For this reason, isolate SY710 was excluded from molecular analysis. A single, faster band was observed in IMA2 which was run as an outgroup species (Fig. 5). Esterase patterns did not show any polymorphism among the different isolates (results not shown).
FIG. 5.
MDH profiles of different G. mosseae isolates and of G. caledonium (IMA2), included for comparison. Lane 1, BEG25; lane 2, IN101C; lane 3, AZ225C; lane 4, SY710; lanes 5 and 6, IMA1; lane 7, IMA2.
Total protein profiles were highly reproducible and the UPGMA dendrogram obtained from the compiled polypeptide data set revealed similarity indices between isolates ranging from 0.60 to 0.79 (Fig. 6). The two American isolates were more similar to IMA1 than to BEG25. Isolate IMA3, included for comparison, belonged to a distinct cluster. The cophenetic correlation coefficient of the dendrogram was 0.95, showing that the tree accurately represented the similarity matrix and bootstrap values were higher than 50%.
FIG. 6.
UPGMA cluster analysis of soluble protein profiles of four geographically different isolates of G. mosseae and of G. coronatum (IMA3), included for comparison.
RFLP-ITS characterization.
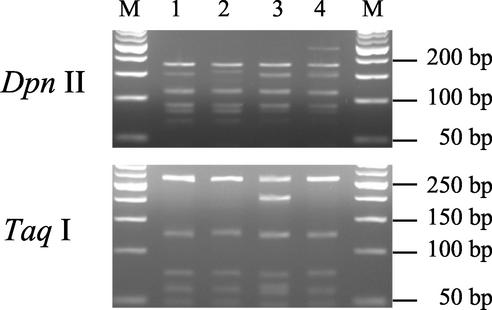
Amplification of the ITS region in the isolates AZ225C, BEG25, IMA1, and IN101C produced uniform fragments of ca. 580 bp. The ITS region from the tested isolates lacked restriction sites for the endonucleases AluI, EcoRI, and HaeIII. HinfI, RsaI, and MspI digestions of ITS did not produce polymorphisms. DpnII gave three different profiles, which made it possible to distinguish BEG25 and IN101C, and TaqI produced two different profiles discriminating IMA1 from the other isolates (Fig. 7). The sizes of fragments obtained from each enzyme and for each pattern are given in Table 3.
FIG. 7.
RFLP patterns produced by restriction digestions of the ITS region of the rDNA of four geographically different isolates of G. mosseae. Isolates were digested with DpnII (top) and with TaqI (bottom). Lane 1, AZ225C; lane 2, BEG25; lane 3, IMA1; lane 4, IN101C; lanes M, size markers.
TABLE 3.
Sizes of fragments obtained from restriction digestion of ITS amplified from the different G. mosseae isolates
| Restriction enzyme | Isolate(s) | Profile fragment sizes (bp) | Total length (bp) |
|---|---|---|---|
| RsaI | AZ225C, BEG25, IMA1, IN101C | 406, 108 | 514 |
| MspI | AZ225C, BEG25, IMA1, IN101C | 272, 179, 137 | 588 |
| HinfI | AZ225C, BEG25, IMA1, IN101C | 352, 190 | 542 |
| DpnII | AZ225C, IMA1 | 183, 156, 115, 94, 80, 68 | 696 |
| DpnII | BEG25 | 183, 165, 115, 94, 80, 68 | 705 |
| DpnII | IN101C | 253, 183, 156, 115, 94, 80, 68 | 949 |
| TaqI | IMA1 | 278, 214, 133 | 625 |
| TaqI | AZ225C, BEG25, IN101C | 278, 133 | 411 |
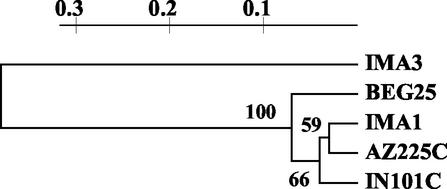
The dendrogram obtained from the RFLP profiles clustered all G. mosseae isolates in a single group, showing appreciable divergence from IMA3 (Fig. 8). As shown in total protein analysis, the two American isolates appeared more similar to IMA1 than to BEG25. Cluster analysis was supported by the cophenetic correlation coefficient (0.99) and bootstrap values (higher than 50%).
FIG. 8.
Dendrogram produced from cluster analysis of RFLP-ITS profiles, pooling data from all restriction enzymes, of four geographically different isolates of G. mosseae and of G. coronatum (IMA3), included for comparison.
DISCUSSION
This study represents the first documentation, to our knowledge, of the occurrence of vegetative incompatibility between different isolates of the AM fungal species G. mosseae. Anastomosis formation in AM fungi had been previously observed by some authors (22, 34, 49), and subsequently detected and quantified both in presymbiotic and symbiotic mycelia (15, 18). Hyphal fusions were reported to occur regularly in mycelia originating from individually germinated spores belonging to the same isolate, in three different Glomus species (15). Here, vegetative compatibility tests showed that six geographically different G. mosseae isolates were capable of self-anastomosing and that anastomosis frequencies ranged between 60 and 85%. Such values are comparable to those reported previously for G. mosseae (51 to 57%) and for self-anastomosing isolates of Rhizoctonia solani (more than 50%) and higher than those reported for Rhizoctonia endophytes from the roots of Pinus silvestris and of the orchid Goodyera repens, ranging from 22 to 50% (24, 45). The ability to produce self-fusions has been reported to affect the fitness of R. solani, with non-self-anastomosing isolates showing lower survival rates and reduced saprophytic and pathogenic ability (24). The self-anastomosing ability shown by the G. mosseae isolates tested supports the hypothesis that this feature may have important fitness consequences, representing a fundamental survival strategy for these obligate biotrophs. Further work will be needed to establish whether spores germinating in the absence of the host can plug into compatible symbiotic mycelia in the soil, thus gaining access to nutrient resources available in the mycorrhizal network (17).
The establishment of protoplasm continuity during self-anastomosis is important for the maintenance of physiological and genetic continuity within each mycelium. The detection of nuclei in hyphal bridges after self-anastomosis in all G. mosseae isolates confirms the occurrence of nuclear migration through the fusion pore developed during hyphal anastomosis (1, 2, 15, 18). Nuclear exchange may have important implications for the biology of these asexual fungi. Recent studies, carried out using fluorescence in situ hybridization, detected genetically different nuclei within individual spores in four species of AM fungi (26, 50). Thus, the occurrence of multiple genomes in these clonal organisms strongly supports the inference that nuclear exchange following anastomosis may represent a fundamental mechanism allowing the maintenance of genetic diversity in the absence of sexual recombination (6, 15, 41).
In this work no successful anastomoses could be detected between germlings of geographically different isolates of G. mosseae. This finding, together with their self-anastomosing capacity, suggests the possibility of assigning each isolate to a distinct VCG (29). Further experiments should be carried out in order to assess the distribution of compatible isolates within the same geographical region and to enhance knowledge on the population structure of AM fungi.
Vegetative incompatibility among G. mosseae isolates was not identified on the basis of the responses usually produced by higher fungi, such as barrage reactions, which were impossible to detect due to the poor growth ability of AM fungal hyphae. Microscopic examinations of hyphal contacts between germlings belonging to different isolates allowed detection of hyphal recognition responses, which were followed by incompatible reactions prior to any anastomosis formation. Tropism before hyphal fusion was previously observed in G. caledonium while monitoring the dynamics of anastomosis formation in living hyphae (19) and was described in Phanerochaete velutina and Stereum spp., which showed different interhyphal attraction distances (1, 2). A previous work reported that no tropism or recognition responses were detected between hyphae of different species or genera of AM fungi (15). Thus, the fact that a hyphal tip was able to change direction and to develop branches prior to any physical contact with a hypha belonging to a conspecific isolate suggests that specific recognition signals may be involved during this stage and that fusion must be highly regulated. However, whether and what specific signals are involved in self-nonself recognition remains to be clarified in these symbiotic fungi.
The development of septa and of empty hyphal tips prior to or following hyphal contacts is the only event observed during incompatible interactions and is comparable to postfusion incompatibility responses described in other fungal species (2, 25, 29, 35). Vegetative incompatibility, preventing hyphal fusion and cytoplasmic and nuclear exchange, has been considered the basis of fungal individualism (36, 54). The strong genetic barriers to hyphal fusions exhibited by G. mosseae isolates of different geographical origins may function to hinder heterokaryon formation between genetically different mycelia, thus permitting the maintenance of the fittest gene combinations. Such barriers may also prevent the exchange of cytoplasm and the spread of harmful genetic elements (20, 29).
The intraspecific genetic diversity of G. mosseae revealed by vegetative compatibility analysis was confirmed by phenotypic and genotypic results: not all the isolates were morphologically identical, although fitting the species description, and both electrophoretic profiles from spore soluble proteins and ITS-RFLP banding patterns clearly distinguished the isolates tested.
No isozyme polymorphism was detected by MDH and EST enzymes, which showed the same electrophoretic pattern in all G. mosseae isolates. By contrast, these enzymes permitted the differentiation of G. mosseae from G. caledonium. Previous studies on the diversity of geographically distinct isolates of glomalean species showed that MDH and EST were able to discriminate isolates belonging to different species (11, 23). Other studies reported that MDH profiles could separate isolates of Gigaspora spp. at the subgeneric level into two different groups, which fit with the analysis of 18S rDNA sequences (5).
Total protein profiles analyzed by SDS-PAGE confirmed vegetative compatibility data, since all the isolates were distinguishable and produced different patterns. Cluster analysis revealed that the two American isolates were more similar to IMA1 than to BEG25 and that similarity indexes between isolates ranged from 60 to 79%. Previous studies showed that different environmental conditions did not affect total protein profiles, which could be utilized to detect inter- and intraspecific differences in AM fungi (4). Other studies reported that interspecific similarity was lower than that observed between conspecific isolates and that G. mosseae was clearly differentiated from other species (4, 11, 55).
In this study all G. mosseae isolates were characterized by unique ITS-RFLP banding patterns, showing that ITS-RFLP analysis may be utilized for organism differentiation at the intraspecific level. This contrasts with previous works reporting that ITS-RFLP profiles, obtained by using a lower number of restriction enzymes, could distinguish AM fungi at the species level (37, 42). Our ITS-RFLP profiles evidenced molecular diversity between the two European isolates IMA1 and BEG25 and demonstrated that IMA1 was more similar to the two American isolates than to BEG25, confirming protein profile analyses. Our findings support results from other works which show variable degrees of genetic differences among G. mosseae isolates BEG25, BEG69, and BEG12 (sharing with IMA1 the same origin as the Rothamsted isolate), depending on which ribosomal region is compared (30, 51). As an example, sequence comparisons of ITS and 5.8S regions showed the diversity of BEG25 and BEG12, which clustered in two groups (30).
In conclusion, our biochemical and molecular findings confirmed the differentiation of G. mosseae isolates obtained by using vegetative compatibility analysis, opening the way to the utilization of vegetative incompatibility as an indicator of genetic diversity between isolates of AM fungi. Moreover, since AM fungi lack a tractable genetic system, vegetative compatibility tests may represent an easy assay for the detection of genetically different mycelia and an additional powerful tool for investigating the population structure and genetics of these obligate symbionts.
Acknowledgments
This work was supported by funds from the University of Pisa and by the C.N.R. (National Research Council, Rome, Italy).
REFERENCES
- 1.Ainsworth, A. M., and A. D. M. Rayner. 1986. Responses of living hyphae associated with self and non-self fusions in the basidiomycete Phanerochaete velutina. J. Gen. Microbiol. 132:191-201. [Google Scholar]
- 2.Ainsworth, A. M., and A. D. M. Rayner. 1989. Hyphal and mycelial responses associated with genetic exchange within and between species of the basidiomycete genus Stereum. J. Gen. Microbiol. 135:1643-1659. [Google Scholar]
- 3.Antoniolli, Z. I., D. P. Schachtman, K. K. Ophel, and S. E. Smith. 2000. Variation in rDNA ITS sequences in Glomus mosseae and Gigaspora margarita spores from a permanent pasture. Mycol. Res. 104:708-715. [Google Scholar]
- 4.Avio, L., and M. Giovannetti. 1998. The protein pattern of spores of arbuscular mycorrhizal fungi: comparison of species, isolates and physiological stages. Mycol. Res. 102:985-990. [Google Scholar]
- 5.Bago, B., S. P. Bentivenga, V. Brenac, J. C. Dodd, Y. Piche, and L. Simon. 1998. Molecular analysis of Gigaspora (Glomales, Gigasporaceae). New Phytol. 139:581-588. [Google Scholar]
- 6.Bever, J. D., and J. Morton. 1999. Heritable variation and mechanisms of inheritance of spore shape within a population of Scutellospora pellucida, an arbuscular mycorrhizal fungus. Am. J. Bot. 86:1209-1216. [PubMed] [Google Scholar]
- 7.Burggraaf, J. P., and J. E. Beringer. 1989. Absence of nuclear DNA synthesis in vesicular-arbuscular mycorrhizal fungi during in vitro development. New Phytol. 111:25-33. [Google Scholar]
- 8.Chillali, M., H. Ideer-Ighili, J. J. Guillaumin, C. Mohammed, B. Lung Escarmant, and B. Botton. 1998. Variation in the ITS and IGS regions of ribosomal DNA among the biological species of European Armillaria. Mycol. Res. 102:533-540. [Google Scholar]
- 9.Cortesi, P., M. G. Milgroom, and M. Bisiach. 1996. Distribution and diversity of vegetative compatibility types in subpopulations of Cryphonectria parasitica in Italy. Mycol. Res. 100:1087-1093. [Google Scholar]
- 10.Dahlberg, A., and J. Stenlid. 1994. Size, distribution and biomass of genets in populations of Suillus bovinus (L.: Fr.) Roussel revealed by somatic incompatibility. New Phytol. 128:225-234. [DOI] [PubMed] [Google Scholar]
- 11.Dodd, J. C., S. Rosendhal, M. Giovannetti, A. Broome, L. Lanfranco, and C. Walker. 1996. Inter- and intraspecific variation within the morphologically-similar arbuscular mycorrhizal fungi Glomus mosseae and Glomus coronatum. New Phytol. 133:113-122. [Google Scholar]
- 12.Douds, D. D., G. Nagahashi, and G. D. Abney. 1996. The differential effects of cell wall associated phenolics, cell walls, and cytosolic phenolics of host and nonhost roots on the growth of two species of AM fungi. New Phytol. 133:289-294. [DOI] [PubMed] [Google Scholar]
- 13.Fries, N. 1987. Somatic incompatibility and field distribution of the ectomycorrhizal fungus Suillus luteus (Boletaceae). New Phytol. 107:735-739. [Google Scholar]
- 14.Gerdemann, J., and J. M. Trappe. 1974. The Endogonaceae of the Pacific Northwest. Mycologia memoires no. 5. Mycological Society of America, New York Botanic Garden, New York, N.Y.
- 15.Giovannetti, M., D. Azzolini, and A. S. Citernesi. 1999. Anastomosis formation and nuclear and protoplasmic exchange in arbuscular mycorrhizal fungi. Appl. Environ. Microbiol. 65:5571-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovannetti, M. 2000. Spore germination and pre-symbiotic mycelial growth, p. 47-68. In Y. Kapulnik and D. D. Douds (ed.), Arbuscular mycorrhizas: physiology and function. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 17.Giovannetti, M. 2001. Survival strategies in arbuscular mycorrhizal symbionts, p. 185-196. In J. Seckbach (ed.), Symbiosis mechanisms and model systems. Kluwer Academic Publisher, Dordrecht, The Netherlands.
- 18.Giovannetti, M., P. Fortuna, A. S. Citernesi, S. Morini, and M. P. Nuti. 2001. The occurrence of anastomosis formation and nuclear exchange in intact arbuscular mycorrhizal networks. New Phytol. 151:717-724. [DOI] [PubMed] [Google Scholar]
- 19.Giovannetti, M., and C. Sbrana. 2001. Self and non-self responses in hyphal tips of arbuscular mycorrhizal fungi, p. 221-231. In A. Geitmann and M. Cresti (ed.), Cell biology of plant and fungal tip growth. NATO science series I: life and behavioural sciences, vol. 328. IOS Press, Amsterdam, The Netherlands.
- 20.Glass, N. L., and G. A. Kuldau. 1992. Mating type and vegetative incompatibility in filamentous ascomycetes. Annu. Rev. Phytopathol. 30:201-224. [DOI] [PubMed] [Google Scholar]
- 21.Glass, N. L., D. J. Jacobson, and P. K. T. Shiu. 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34:165-186. [DOI] [PubMed] [Google Scholar]
- 22.Godfrey, R. M. 1957. Studies on British species of Endogone. III. Germination of spores. Trans. Br. Mycol. Soc. 40:203-210. [Google Scholar]
- 23.Hepper, C. M., R. Sen, C. Azcon-Aguilar, and C. Grace. 1988. Variation in certain isozymes amongst different geographical isolates of the vesicular-arbuscular mycorrhizal fungi Glomus clarum, Glomus monosporum and Glomus mosseae. Soil Biol. Biochem. 20:51-59. [Google Scholar]
- 24.Hyakumachi, M., and T. Ui. 1987. Non-self-anastomosing isolates of Rhizoctonia solani obtained from fields of sugarbeet monoculture. Trans. Br. Mycol. Soc. 89:155-159. [Google Scholar]
- 25.Jacobson, D. J., K. Beurkens, and K. L. Klomparens. 1998. Microscopic and ultrastructural examination of vegetative incompatibility in partial diploids heterozygous at het loci in Neurospora crassa. Fung. Genet. Biol. 23:45-56. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn, G., M. Hijri, and I. R. Sanders. 2001. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature 414:745-748. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lanfranco, L., M. Delpero, and P. Bonfante. 1999. Intrasporal variability of ribosomal sequences in the endomycorrhizal fungus Gigaspora margarita. Mol. Ecol. 8:37-45. [DOI] [PubMed] [Google Scholar]
- 29.Leslie, J. F. 1993. Fungal vegetative compatibility. Annu. Rev. Phytopathol. 31:127-150. [DOI] [PubMed] [Google Scholar]
- 30.Lloyd-McGilp, S. A., S. M. Chambers, J. C. Dodd, A. H. Fitter, C. Walker, and J. P. W. Young. 1996. Diversity of the ribosomal internal transcribed spacers within and among isolates of Glomus mosseae and related mycorrhizal fungi. New Phytol. 133:103-111. [Google Scholar]
- 31.Logi, C., C. Sbrana, and M. Giovannetti. 1998. Cellular events involved in survival of individual arbuscular mycorrhizal symbionts growing in the absence of the host. Appl. Environ. Microbiol. 64:3473-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mes, J. J., E. A. Weststeijn, F. Herlaar, J. J. M. Lambalk, J. Wijbrandi, M. A. Haring, and B. J. C. Cornelissen. 1999. Biological and molecular characterization of Fusarium oxysporum f.sp. lycopersici divides race 1 isolates into separate virulence groups. Phytopathology 89:156-160. [DOI] [PubMed] [Google Scholar]
- 33.Milgroom, M. G., and P. Cortesi. 1999. Analysis of population structure of the chestnut blight fungus based on vegetative incompatibility genotypes. Proc. Natl. Acad. Sci. USA 96:10518-10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosse, B. 1959. The regular germination of resting spores and some observations on the growth requirements of an Endogone sp. causing vesicular-arbuscular mycorrhiza. Trans. Br. Mycol. Soc. 42:273-286. [Google Scholar]
- 35.Newhouse, J. R., and W. L. MacDonald. 1991. The ultrastructure of hyphal anastomoses between vegetatively compatible and incompatible virulent and hypovirulent strains of Cryphonectria parasitica. Can. J. Bot. 69:602-614. [Google Scholar]
- 36.Rayner, A. D. M. 1991. The challenge of the individualistic mycelium. Mycologia 83:48-71. [Google Scholar]
- 37.Redecker, D., H. Thierfelder, C. Walker, and D. Werner. 1997. Restriction analysis of PCR-amplified internal transcribed spacers of ribosomal DNA as a tool for species identification in different genera of the order Glomales. Appl. Environ. Microbiol. 63:1756-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohlf, F. J., and R. R. Sokal. 1981. Comparing numerical taxonomic studies. Syst. Zool. 30:459-490. [Google Scholar]
- 39.Rosendhal, S., and R. Sen. 1992. Isozyme analysis of mycorrhizal fungi and their mycorrhiza. Methods Microbiol. 24:169-194. [Google Scholar]
- 40.Rosendhal, S., and J. W. Taylor. 1997. Development of multiple genetic markers for studies of genetic variation in arbuscular mycorrhizal fungi using AFLP. Mol. Ecol. 6:821-829. [Google Scholar]
- 41.Sanders, I. 1999. No sex please, we are fungi. Nature 399:737-739. [DOI] [PubMed] [Google Scholar]
- 42.Sanders, I. R., M. Alt, K. Groppe, T. Boller, and A. Wiemken. 1995. Identification of ribosomal DNA polymorphisms among and within spores of the Glomales: application to studies on the genetic diversity of arbuscular mycorrhizal fungal communities. New Phytol. 130:419-427. [Google Scholar]
- 43.Sbrana, C., L. Avio, and M. Giovannetti. 1995. The occurrence of calcofluor and lectin binding polysaccharides in the outer wall of AM fungal spores. Mycol. Res. 99:1249-1252. [Google Scholar]
- 44.Sen, R. 1990. Intraspecific variation in two species of Suillus from Scots pine (Pinus sylvestris L.) forests based on somatic incompatibility and isozyme analyses. New Phytol. 114:607-616. [Google Scholar]
- 45.Sen, R., A. M. Hietala, and C. D. Zelmer. 1999. Common anastomosis and internal transcribed spacer RFLP groupings in binucleate Rhizoctonia isolates representing root endophytes of Pinus sylvestris, Ceratorhiza spp. from orchid mycorrhizas and a phytopathogenic anastomosis group. New Phytol. 144:331-341. [Google Scholar]
- 46.Smith, S. E., and V. Gianinazzi-Pearson. 1990. Phosphate uptake and arbuscular activity in mycorrhizal Allium cepa L.: effects of photon irradiance and phosphate nutrition. Aust. J. Plant Physiol. 17:177-188. [Google Scholar]
- 47.Smith, S. E., and D. J. Read. 1997. Mycorrhizal symbiosis. Academic Press, London, United Kingdom.
- 48.Stenlid, J., and R. Vasiliauskas. 1998. Genetic diversity within and among vegetative compatibility groups of Stereum sanguinolentum determined by arbitrary primed PCR. Mol. Ecol. 7:1265-1274. [Google Scholar]
- 49.Tommerup, I. C. 1988. The vesicular-arbuscular mycorrhizas. Adv. Plant Pathol. 6:81-91. [Google Scholar]
- 50.Trouvelot, S., D. van Tuinen, M. Hijiri, and V. Gianinazzi-Pearson. 1999. Visualization of ribosomal DNA loci in spore interphasic nuclei of glomalean fungi by fluorescence in situ hybridization. Mycorrhiza 8:201-206. [Google Scholar]
- 51.Vandenkoornhuyse, P., and Leyval, C. 1998. SSU rDNA sequencing and PCR-fingerprinting reveal genetic variation within Glomus mosseae. Mycologia 90:791-797. [Google Scholar]
- 52.Van de Peer, Y., and R. De Wachter. 1994. A software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 53.Viera, A., and M. G. Glenn. 1990. DNA content of vesicular-arbuscular mycorrhizal fungal spores. Mycologia 82:263-267. [Google Scholar]
- 54.Worrall, J. J. 1997. Somatic incompatibility in basidiomycetes. Mycologia 89:24-36. [Google Scholar]
- 55.Xavier, L. J. C., I. J. Xavier, and J. J. Germida. 2000. Potential of spore protein profiles as identification tools for arbuscular mycorrhizal fungi. Mycologia 82:1210-1213. [Google Scholar]