Abstract
Intergeneric conjugal transfer of plasmid DNA from Escherichia coli to Streptomyces circumvents problems such as host-controlled restriction and instability of foreign DNA during the transformation of Streptomyces protoplasts. The anthracycline antibiotic-producing strains Streptomyces peucetius and Streptomyces sp. strain C5 were transformed using E. coli ET12567(pUZ8002) as a conjugal donor. When this donor species, carrying pSET152, was mated with Streptomyces strains, the resident plasmid was mobilized to the recipient and the transferred DNA was also integrated into the recipient chromosome. Analysis of the exconjugants showed stable integration of the plasmid at a single chromosomal site (attB) of the Streptomyces genome. The DNA sequence of the chromosomal integration site was determined and shown to be conserved. However, the core sequence, where the crossover presumably occurred in C5 and S. peucetius, is TTC. These results also showed that the φC31 integrative recombination is active and the phage attP site is functional in S. peucetius as well as in C5. The efficiency and specificity of φC31-mediated site-specific integration of the plasmid in the presence of a 3.7-kb homologous DNA sequence indicates that integrative recombination is preferred under these conditions. The integration of plasmid DNA did not affect antibiotic biosynthesis or biosynthesis of essential amino acids. Integration of a single copy of a mutant chiC into the wild-type S. peucetius chromosome led to the production of 30-fold more chitinase.
Streptomyces spp. are gram-positive spore-forming soil bacteria, and they synthesize a wide array of antibiotics and other secondary metabolites. Genetic approaches to improve secondary-metabolite production in antibiotic producer strains were hampered by restriction barriers, the absence of efficient gene transfer systems in industrial strains, and a lack of suitable cloning vectors (4). To circumvent these problems, there has been considerable interest in the use of intergeneric conjugation as a means of plasmid transfer, using Escherichia coli as the donor. This technique allows one to construct and manipulate recombinant plasmids in E. coli and subsequently transfer them to Streptomyces. This mode of gene transfer has been efficient even in recipient strains containing restriction systems (5, 33).
Intergeneric conjugation between E. coli and Streptomyces was first reported in 1989 (18). Since then, the process has been demonstrated to work in several Streptomyces species, such as Streptomyces fradiae, Streptomyces ambofaciens (5), Streptomyces coelicolor, Streptomyces lividans (8), and Streptomyces nanchangensis (28); in Saccharopolyspora spinosa (17); and in various strains of actinomycetales (32). In all these cases, the donor species was E. coli. The conjugative functions of plasmid RP4, (5, 10, 16, 18, 21, 26, 32), the RP1 derivative pMB307 (8), and the RK2 derivative pUZ8002 (23) were used to mobilize the resident plasmids. Several cloning vectors which could be transferred from E. coli to Streptomyces spp. by conjugation have been constructed. These vectors are nonreplicative in Streptomyces but could integrate into the chromosomal attachment site of bacteriophage φC31 (5, 9, 29, 33), plasmid pSAM2 (12, 26), or minicircle DNA (21).
Stable integration of cloned genes into the bacterial chromosome minimizes the problems associated with the presence of multiple copies of foreign genes, as well as the instability of the recombinants (5). Recombinants containing a single copy of an antibiotic gene cluster inserted at the φC31 attachment site produced amounts of tetracyclic macrolide (in S. spinosa [16]) and glycopeptide (in Streptomyces toyocaensis [17]) comparable to those of the wild type without any deleterious effect on the biosynthesis of secondary metabolites. Integration of a 12-kb fragment harboring the megosamine pathway resulted in efficient conversion of erythromycin to megalomicin in Saccharopolyspora erythraea (34).
Streptomyces peucetius and Streptomyces sp. strain C5 produce the polyketide antibiotics daunorubicin and doxorubicin. These antibiotics are of clinical interest because of their antineoplastic activities (27). The aim of this study was to demonstrate intergeneric conjugation between E. coli and S. peucetius and Streptomyces sp. strain C5 and expression of the integrated foreign genes in exconjugants. The integration site was cloned and sequenced from several exconjugants, and the results show that there is a single integration site for C5 and S. peucetius. Comparison of nucleotide sequences with databases revealed that the nucleotide sequence of the integration locus is conserved among the Streptomyces strains examined. However, the crossover region has a trinucleotide, TTG, in S. coelicolor and S. lividans, but S. peucetius and strain C5 have TTC, and the sequences of C5 and S. peucetius spanning the crossover region revealed 88 and 93% identity with the gene that encodes a chromosomal condensation protein of S. coelicolor. The efficiency of φC31-directed site-specific recombination was not affected by the presence of homologous genomic DNA sequences present in the plasmid vector. Furthermore, the integration of foreign DNA did not affect growth, sporulation, antibiotic production, or amino acid biosynthesis, indicating that the location of the integration site is “neutral.” Using this system, we examined the integration and expression of an extra copy of the chitinase gene in S. peucetius.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain DH5α was used as the general cloning host, and E.coli strain ET12567(pUZ8002) was used as the donor in intergeneric conjugation. ET12567 is a methylation-defective strain (dam-13::Tn9 dcm-6 hsdM Cmr) (provided by Iain S. Hunter, University of Strathclyde). Plasmid pUZ8002 is a derivative of RK2 with a mutation in oriT. The efficiency of conjugal transfer of this plasmid was 1,000-fold less than that of RK2, but it is capable of mobilizing other plasmids efficiently (23). Streptomyces sp. strain C5 and S. peucetius were provided by W. Strohl (Merck Research Laboratorieis, Rahway, N.J.). SPVI was isolated in our laboratory by N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis of S. peucetius (30). S. peucetius ATCC 29050 was from the American Type Culture Collection. The integrative plasmid pSET152 (provided by Iain S. Hunter; accession number AJ414670) carries φC31 int and att functions and an apramycin resistance gene for selection in Streptomyces and E. coli (5).
Media and culture conditions.
E. coli strains were grown in Luria broth (LB) supplemented with antibiotics. Apramycin (50 μg/ml; Sigma Chemical Co., St. Louis, Mo.), chloramphenicol (25 μg/ml), and kanamycin (50 μg/ml) were added to growth media as required. Soybean mannitol agar was used for the preparation of spore stocks of C5 and for maintaining S. peucetius. Tryptone soya (TS) broth supplemented with 5 μg of apramycin/ml was used to grow exconjugants for the preparation of genomic DNA. Soybean mannitol agar supplemented with 50 μg of apramycin/ml was used to maintain the exconjugants. For the chitinase assay, S. peucetius cultures were grown in NDYE (6) for 48 h, and the mycelia were sedimented at 2,000 × g for 10 min at 4°C. The packed mycelial volume was measured, and the mycelia were used as the inoculum for 50 ml of nitrate defined medium containing 5 mg of insoluble chitin/ml as the sole carbon source (NDMC) (30).
Intergeneric conjugation.
Intergeneric conjugation between E. coli and Streptomyces was performed as described previously (8) with minor modifications. E. coli ET12567(pUZ8002/pSET152) was grown to an absorbance of 0.4 at 600 nm (A600). The cells were pelleted by centrifugation, washed twice in an equal volume of LB, pelleted again, and finally resuspended in 1/10 volume of LB. Aliquots of Streptomyces sp. strain C5 spore suspension stored at −20°C were used as recipients. Spores (∼108) were washed in 2× yeast extract tryptone medium, resuspended in 500 μl of 2× yeast extract tryptone medium, and incubated at 50°C for 10 min to induce germination. Donor cells (500 μl; 108 cells) were added to the treated spores, the mixture was pelleted by centrifugation, and finally the pellet was resuspended in the residual liquid and the undiluted mixture was plated. The mating mixture was spread on soybean mannitol agar plates containing 10 mM MgCl2 and incubated at 30°C for 18 h. The plates were overlaid with 1 ml of water containing 500 μg of nalidixic acid and 1 mg of apramycin. The plates were incubated further for 5 days at 30°C, and the exconjugants were counted; cultures prepared from single spores were used for further studies. A control experiment was performed as described above but without the addition of E. coli donor cells. The viable count of the donor culture was determined by spreading the cells on LB agar plates supplemented with appropriate antibiotics.
Mycelial fragments were used as recipients in the case of S. peucetius, since the strain sporulated poorly. The mycelial fragments were prepared for mating as described earlier (5) with the following modifications. One milliliter of S. peucetius mycelial culture was diluted with 9 ml of TS broth and cultured for 18 h at 30°C in a shaker. The culture was homogenized (2), and 2 ml was mixed with 18 ml of fresh TS broth and grown for 16 h at 30°C. This culture was again homogenized and sonicated with the following settings: 36% amplitude; temperature, 28°C; pulse on for 3 s, pulse off for 4 s for two cycles using a Sonics-Vibracell. One milliliter of the culture was diluted with 9 ml of TS broth, and the suspension was incubated aerobically at 30°C for 3 h. The mycelia were recovered by centrifugation, washed once in TS broth, and resuspended in 2 ml of TS broth. The donor cells (100 μl; 108 cells) and 100 μl of mycelial fragments were mixed. The undiluted conjugation mixture was spread as described above. After two cycles of single-colony purification on selective plates, the exconjugants were used for further experiments. The viable count of the recipient was determined by spreading the mycelial fragments on soybean mannitol agar plates.
DNA techniques.
DNA cloning was carried out essentially as described earlier (24). Restriction endonucleases were obtained from Promega or Boehringer Mannheim Biochemicals and were used according to the specifications of the manufacturers. Total genomic DNA was isolated from Streptomyces spp. as described earlier (11). The DNA was transferred to BIODYNE nylon membranes (GIBCO BRL) using a vacuum blotting system (Hoefer, San Francisco, Calif.). The hybridization conditions and subsequent detection were in accordance with the manufacturer's instructions (GIBCO BRL). Preparation of the DNA probe was carried out using the Prime-a-Gene kit (Promega) following the manufacturer's instructions.
Plasmid construction.
A 3.7-kb BamHI DNA fragment from S. peucetius carrying the antibiotic biosynthetic pathway gene cluster dnrUVJI was ligated to the BamHI-digested pSET152 plasmid to generate plasmid pPD1. A 2.7-kb BamHI/EcoRI fragment carrying the S. peucetius chitinase gene (chiC) from plasmid pSPCHIC (K. S. Vetrivel, unpublished results) and a 2.7-kb BamHI/EcoRI fragment carrying the chitinase gene (chiC) from a chitinase-overproducing S. peucetius mutant, SPVI (31), were cloned into BamHI/EcoRI-digested pSET152 and named pPVD1 and pPVD2, respectively. The construction of other plasmids is described below.
Cloning the hybrid integration site.
The presence of a single BamHI site in pSET152 plasmid DNA allowed the cloning of one of the phage-host hybrid integration sites as described below. One microgram of genomic DNA from an exconjugant was digested with BamHI, and the digested DNA was purified and self-ligated with T4 DNA ligase. The ligated total DNA was used for transforming E. coli DH5α. Apramycin-resistant transformants were selected, and the plasmid DNA was isolated. Restriction analysis showed the presence of vector as well as chromosomal DNAs.
Nucleotide sequence of the insertion site.
Plasmids pPD4.1 and pPD6.1 carrying the hybrid attachment sites cloned from C5 and S. peucetius exconjugants were digested with SphI, and the resulting fragments were subcloned into M13mp18 digested with SphI. Clones carrying inserts in two different orientations were identified using C-Test analysis as described previously (7). Briefly, single-stranded DNAs from the recombinant phages were mixed with 1% sodium dodecyl sulfate (SDS) and loading dye, incubated at 65°C for 1 h, and then allowed to cool to room temperature. Clones carrying single-stranded DNAs with different orientations anneal together and form double-stranded DNA under these conditions, and the mobility of such DNA was retarded on agarose gels. Dideoxy sequencing (25) was performed using a T7 sequenase version 2.0 DNA-sequencing kit (Amersham-Pharmacia) as described by the manufacturer. Double-strand sequencing was carried out using an automated DNA sequencer.
Determination of DNA sequence.
The DNA sequence was compiled using the Genetics Computer Group program, and sequence comparisons were performed using the BLAST program from the National Center for Biotechnology Information World Wide Web server (1). Multiple-sequence analysis was performed using the Genetics Computer Group program and edited using the GeneDoc program (http://www.cis.com/∼ketchup/genedoc.shtml).
Chitinase assay.
For chitinase production, Streptomyces cultures were grown in NDYE for 48 h, and the mycelia were pelleted at 2,000 × g for 10 min at 4°C. Equal amounts (wet weight) of packed mycelia were used as the inoculum for 50 ml of NDMC. For enzyme assays, samples were drawn every 24 h, the mycelia were pelleted (10,000 × g; 10 min), and the supernatant was used for the enzyme assay.
The assay for chitinase was carried out as described previously (31) using the fluorogenic substrate 4-methylumbelliferyl-N,N′,N"-triacetyl chitotriose (Sigma Chemical Co.) (19). The amount of 4-methylumbelliferone liberated was measured using a fluorimeter (Hoefer) with an excitation wavelength of 365 nm and an emission wavelength of 460 nm. One unit of chitinase activity is defined as the amount of enzyme required to liberate 1 μmol of 4-methylumbelliferone from the substrate in 1 min at 37°C.
Total protein assay.
Protein concentrations were determined according to the method of Lowry et al. (15) using bovine serum albumin as the standard.
Electrophoresis.
SDS-polyacrylamide gel electrophoresis (PAGE) (12% [wt/vol]) was performed as described earlier (13). Proteins in the gels were fixed with formaldehyde-methanol-water (40:33:27) and silver stained as described previously (22). The molecular masses of the proteins were estimated using SDS-PAGE high-range protein molecular mass standards (Amersham) as markers.
Nucleotide sequence accession number.
The nucleotide sequences of Streptomyces sp. strain C5 and S. peucetius attR have been submitted to the GenBank database under accession numbers AF327404 and AF327405, respectively.
RESULTS
Conjugal transfer of the plasmid pSET152 and φC31-directed site-specific recombination in S. peucetius and Streptomyces sp. strain C5.
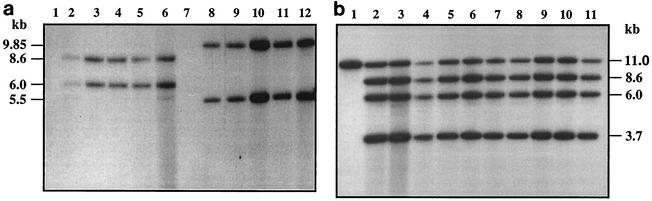
Under the mating conditions used in this study, 1.1 × 10−5 exconjugants per recipient spore could be obtained in the case of Streptomyces sp. strain C5. However, pSET152 could not be transferred by intergeneric conjugation using spores of another isolate of S. peucetius, ATCC 29050, perhaps due to the heat sensitivity of the spores, since the spores were incubated at 50°C to induce germination (S. Paranthaman, unpublished results). Since most of our studies were done using S. peucetius, the mycelial fragments were used as recipients, and in that case, the frequency of exconjugants was 1.5 × 10−4 per recipient cell. The integrative plasmid pSET152 does not replicate in Streptomyces; hence, stable exconjugants can be obtained only if the plasmid is integrated into the chromosome. In order to identify the chromosomal integration site in these recipient strains, chromosomal DNA was extracted from different exconjugants and digested with BamHI, blotted onto membranes, and probed with labeled pSET152 DNA digested with BamHI. Since the plasmid pSET152 has a single BamHI site, DNA from exconjugants are expected to show hybridization of two fragments to the probe. The data in Fig. 1a show that all five of the exconjugants analyzed had two fragments hybridizing to the probe. The fragment sizes were 8.6 and 6.0 kb (Fig. 1a, lanes 2 to 6) in the case of Streptomyces sp. strain C5 and 9.85 and 5.5 kb (lanes 8 to 12) in the case of S. peucetius. In the case of untransformed strains (lanes 1 and 7), the probe did not hybridize to any DNA fragment. The identical pattern of hybridization of BamHI digests observed in all the exconjugants of the strains analyzed implies that the plasmid presumably was integrated at the same locus in all the exconjugants in both strains. The exconjugants containing an insert were stable even after repeated subculturing (up to 50 times) of the mycelia in the absence of antibiotic selection.
FIG. 1.
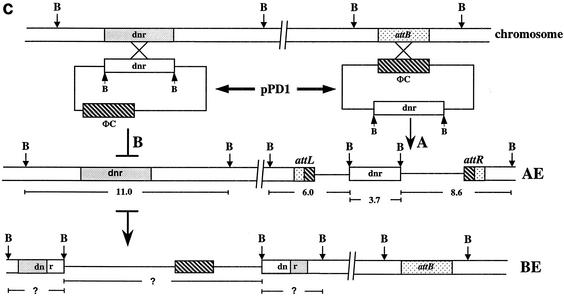
(a) Southern blot of BamHI-digested total DNA. Lanes 1 and 7, untransformed Streptomyces sp. strain C5 and S. peucetius; lanes 2 to 6, apramycin-resistant exconjugants of C5; lanes 8 to 12, apramycin-resistant exconjugants of S. peucetius. DNA was blotted and hybridized with radiolabeled pSET152. (b) Hybridization of 32P-labeled pPD1 (pSET152:dnrUVJI) to BamHI-digested total DNA from untransformed Streptomyces sp. strain C5 (lane 1) and apramycin-resistant exconjugants (lanes 2 to 11). (c) Schematic representation of the integration of plasmid pPD1 in the chromosome of Streptomyces sp. strain C5 by site-specific (AE) and homologous (BE) recombination. Plasmid pPD1carries the φC31 int-att (hatched boxes) and the dnr locus (open boxes) from S. peucetius. The resident copy of the dnr locus (shaded boxes), as well as the attB region (stippled boxes), present in the chromosome of C5 are also shown. The attachment sites attL and attR are the left and right hybrid sites. B, restriction site for BamHI.
Allele replacement by homologous recombination is inefficient in plasmids carrying the φC31 att site.
In order to discover whether integration could occur by homologous recombination in C5 and S. peucetius in the presence of φC31 int and att, a 3.7-kb fragment carrying dnrUVJI genes from the S. peucetius anthracycline antibiotic biosynthetic pathway was cloned into pSET152. This recombinant plasmid, pPD1, was mobilized by intergeneric conjugation from E. coli ET12567(pUZ8002/pPD1) to Streptomyces sp. strain C5 and S. peucetius. The frequency of exconjugant formation with pPD1 was comparable to those obtained with the pSET152 vector alone.
Total genomic DNA was extracted from 10 different C5 exconjugants, cleaved with BamHI, blotted onto membranes, and probed with labeled pPD1. A single BamHI fragment of 11 kb from untransformed cells (Fig. 1b, lane 1) hybridized to the probe, whereas in all 10 exconjugants, the probe hybridized to four fragments (lanes 2 to 11). The 11-kb fragment that hybridized in the untransformed sample was also present in all 10 exconjugants analyzed. If the plasmid integration occurred by homologous recombination, then the 11-kb fragment would have been disrupted by the insertion of the plasmid sequence. However, the 11-kb fragment was unaltered, indicating that the plasmid pPD1 failed to integrate at the dnr cluster. The probe also hybridized to three other DNA fragments of 8.6, 6.0, and 3.7 kb. The deduced restriction map of this location is schematically represented in recombinant type AE in Fig. 1c. The other recombinant type, BE, in Fig. 1c represents the organization of the locus with the plasmid pPD1 integrated by homologous recombination. In that situation, the 11-kb fragment would be disrupted and subsequent BamHI digestion would release three fragments of unknown sizes. The Southern analysis in Fig. 1b clearly shows that all 10 exconjugants obtained in this experiment were obtained after site-specific integration of the plasmid pPD1, and this is represented in Fig. 1c.
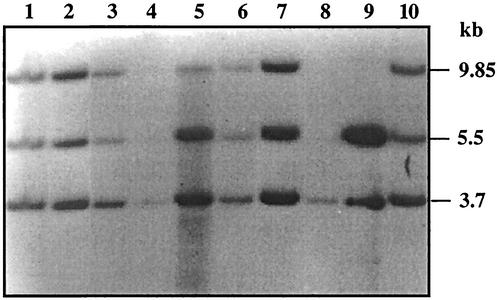
A similar experiment was also carried out in S. peucetius. The total genomic DNA was extracted from the exconjugants, digested with BamHI, blotted onto membranes, and probed with pPD1. In the case of S. peucetius, the results (Fig. 2) indicate that the probe hybridized to 9.85-, 5.5-, and 3.7-kb fragments (Fig. 2, lanes 1 to 7 and 10), whereas it hybridized to a single 3.7-kb fragment (lane 8) in the untransformed strain. The size of the BamHI fragment in which dnrUVJI is located in the S. peucetius chromosome is 3.7 kb, whereas it is 11 kb in C5 (D. Arun and K. Dharmalingam, unpublished data). The 3.7-kb fragment hybridized in the untransformed strain was also present in all of the exconjugants analyzed, indicating that the plasmid pPD1 integrated at a different site in the genome. The 9.85- and 5.5-kb fragments were the junction fragments. This clearly shows that the plasmid pPD1 integrated by site-specific recombination in the genome of S. peucetius.
FIG. 2.
Southern blot of BamHI-digested total DNA. Lane 8, untransformed S. peucetius; lanes 1 to 7 and 10, exconjugants; lane 9, pPD1 plasmid DNA. The probe used was 32P-labeled pPD1 DNA.
DNA sequence of the integration site.
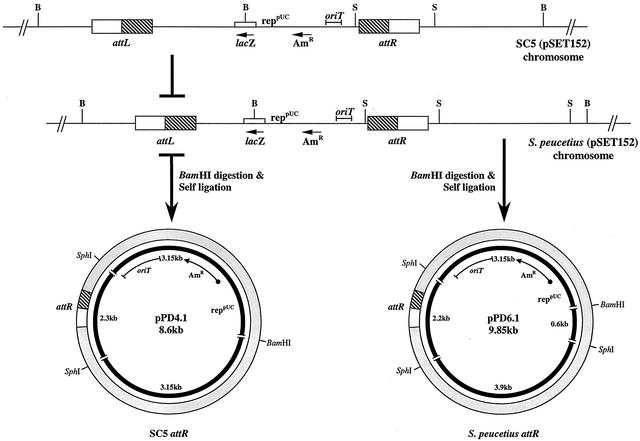
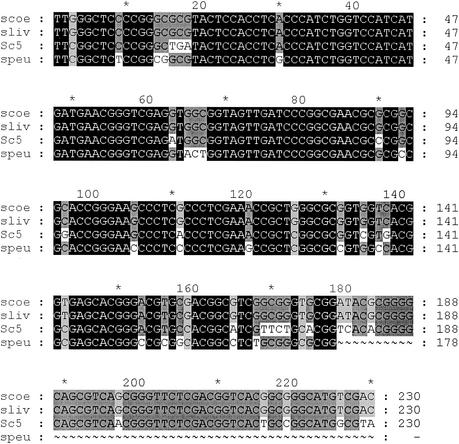
The hybrid attachment site of C5 was cloned and sequenced as described in Materials and Methods. The recombinant plasmid pPD4.1 carries one of the hybrid sites (attR). The other hybrid site (attL) could not be cloned, since the clones carrying this region did not have an origin of replication in the self-ligated DNA. As schematically represented in Fig. 3, BamHI/SphI double digests of the recombinant plasmid pPD4.1 released a 2.3-kb SphI fragment. This corresponds to the 2.3-kb SphI DNA fragment containing part of the plasmid DNA and a portion of C5 genomic DNA including the attR region. Two recombinant plasmids were also generated from S. peucetius exconjugants (pPD6.1 and pPD6.2; 9.85 kb). Both plasmids released a 2.2-kb SphI fragment carrying part of the vector sequence, a portion of the S. peucetius genomic DNA sequence, and a 3.9-kb S. peucetius genomic DNA sequence. The results were confirmed by restriction analysis. The release of a single SphI fragment of the same size, 2.3 kb, from each of three clones of C5 (pDP4.1, pDP4.2, and pDP4.3) confirms that the plasmid pSET152 integrated at a single locus in the genome of Streptomyces sp. strain C5 and presumably in the same orientation. In S. peucetius, the release of 2.2- and 3.9-kb fragments after SphI digestion of the two clones confirms the integration of the plasmid at a single site and in the same orientation. The 2.3- and 2.2-kb SphI fragments were cloned in M13mp18 in both orientations, and the DNA sequence was determined using a forward primer (a 23-mer from GIBCO BRL), as well as internal primers synthesized using the sequence data. Double-stranded DNA sequencing was carried out using an automated sequencer. Comparison of the DNA sequences from C5 with databases revealed 89% identity in a 227-bp overlap with attR junction sequences of S. coelicolor, 88.5% identity in a 227-bp overlap with attR junction sequences of S. lividans, and 88% identity in a 221-bp overlap with an S. coelicolor gene that encodes a chromosomal condensation protein. The sequences from S. peucetius exconjugants revealed 87.4% identity in a 175-bp overlap with attR junction sequences of S. lividans and S. coelicolor and 93% identity in a 54-bp overlap with an S. coelicolor gene that encodes a chromosomal condensation protein. The core sequence, TTG, which is conserved in all att sites of Streptomyces, was found to be TTC in C5 and S. peucetius. Figure 4 shows the multiple-sequence alignment of Streptomyces sp. strain C5 and S. peucetius attR sites with S. lividans and S. coelicolor attR sequences.
FIG. 3.
Schematic representation of cloning of the attR locus from the exconjugants of C5 and S. peucetius. The recombinant plasmids obtained by BamHI-digested total DNA from the exconjugant were self-ligated and transformed into E. coli. The resulting transformants were selected with apramycin. The restriction sites for the BamHI (B) and SphI (S) enzymes are shown. AmR, apramycin resistance. Open box, chromosomal DNA; hatched box, phage DNA.
FIG. 4.
Alignment of attR nucleotide sequences of Streptomyces sp. strain C5 (Sc5) and S. peucetius (speu) with attR sequences of S. lividans (sliv) and S. coelicolor (scoe). Solid boxes, identical sequences; dark shading, 75% conserved; light shading, 50% conserved.
Site-specific integration does not affect the essential functions of Streptomyces.
In order to determine whether the site-specific integration of the plasmid pSET152 affects essential functions of Streptomyces sp. strain C5 and S. peucetius, the spores of the C5 exconjugant and fragmented mycelia of S. peucetius exconjugants were serially diluted and plated on minimal medium (11). The exconjugants grew well in the minimal medium. This indicates that the integration of the plasmid pSET152 did not affect the genes involved in amino acid biosynthesis, as well as antibiotic biosynthesis (data not shown). In addition, freezing, thawing, and germination of the spores did not alter the stability of the recombinant plasmid or its expression.
Integration of one extra copy of chiC gene at φC31 site leads to overproduction of chitinase.
Recently, a daunorubicin pathway-defective mutant (SPVI) of S. peucetius, which was found to overproduce an extra cellular chitinase (30), was isolated. The chitinase gene, chiC, from SPVI was cloned and sequenced (31).
In order to demonstrate the usefulness of this system, a 2.7-kb BamHI/EcoRI fragment harboring SPVI or the wild-type chitinase gene chiC was cloned in pSET152. Plasmids pPVD1 and pPVD2 carry the cloned intact chiC gene of the wild type and SPVI, respectively. E. coli harboring these plasmids was mated with S. peucetius to obtain the strains SPD1 and SPD2. The stable integration of the plasmids pPVD1 and pPVD2 in the genome of S. peucetius was confirmed by Southern analysis (data not shown).
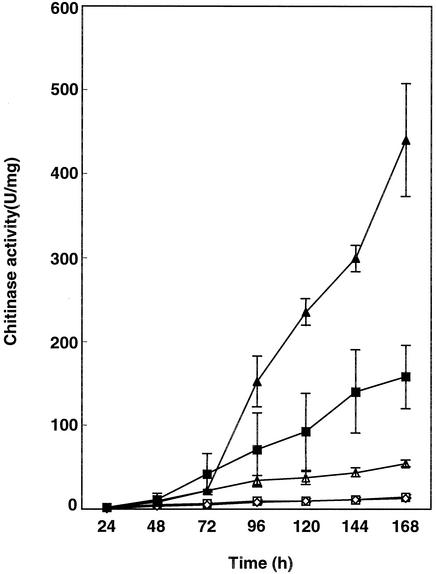
S. peucetius SPVI and the exconjugants SPD1 and SPD2 were grown in NDMC (in triplicate). The inoculum sizes used in these experiments for the parental strain and the exconjugants were identical. Samples were taken every 24 h, and the chitinase assay was carried out using the culture supernatant. At the peak period (Fig. 5), the wild type produced very low levels of chitinase, whereas SPVI showed a >11-fold-higher level of chitinase. The exconjugant SPD2 produced 2.8-fold more chitinase than SPVI and 30-fold more chitinase than the wild type and the exconjugant carrying pSET152. The exconjugant SPD1 carrying wild-type chiC produced 3.8-fold more chitinase than the wild type and 8-fold less than SPVI. The time course study of extracellular chitinase production in SPD2 showed the onset of enzyme production after 72 h.
FIG. 5.
Time course analysis of chitinase production in S. peucetius, SPVI, and exconjugants SPD1 and SPD2 in NDMC. □, S. peucetius; ▵, S. peucetius(pPVD1) (SPD1); ▪, SPVI; ▴, S. peucetius(pPVD2) (SPD2); ⋄, S. peucetius(pSET152). The error bars indicate standard deviations.
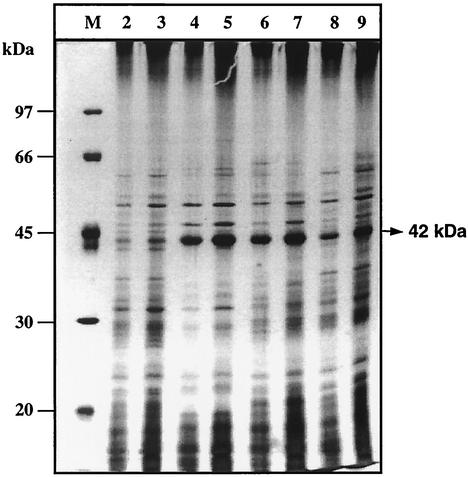
Total extracellular protein profiles of S. peucetius, SPVI, SPD1, and SPD2 were also analyzed by SDS-PAGE (Fig. 6). A protein with a molecular mass of 42 kDa was induced at high levels in SPVI and SPD2 compared to S. peucetius. In the case of the exconjugant SPD1, more of the 42-kDa protein than the wild type was produced, as expected. This increase in the 42-kDa protein correlates well with the chitinase activity. Western analysis using antibodies raised against chitinase confirmed that the 42-kDa protein is chitinase (data not shown).
FIG. 6.
Analysis of extracellular proteins of culture supernatants. Cultures were grown in NDMC for 168 h. SDS-PAGE analysis was performed as described in Materials and Methods. Lane M, molecular mass marker. Five and 10 μg of total extracellular proteins were analyzed. Lanes 2 and 3, S. peucetius; lanes 4 and 5, SPVI; lanes 6 and 7, SPD2; lanes 8 and 9, SPD1.
DISCUSSION
Expression of multiple copies of cloned genes leads to unexpected variations in secondary-metabolite production (3, 4, 17). Moreover, high-copy-number vectors such as pIJ702 were shown to be structurally unstable, and recombinant pIJ702 plasmids frequently showed deletions of cloned inserts (14, 20). One of the best ways to stably clone a single copy of a gene is to integrate the gene of interest in the chromosome itself. With this goal, the plasmid pSET152 was mobilized from E. coli ET12567(pUZ8002) into Streptomyces sp. strain C5 and S. peucetius. Exconjugants were obtained at a frequency of 1.1 × 10−5 and 1.5 × 10−4 per recipient for Streptomyces sp. strain C5 (spores) and S. peucetius (mycelial fragments), respectively. The 10-fold increase in the frequency of exconjugants in the case of S. peucetius mycelial fragments was reproducible. It was shown (16) earlier that S. spinosa mycelial fragments were better recipients than spores for conjugation with E. coli. Unlike the earlier reports, which showed a high frequency of exconjugants, in our studies the frequency of exconjugant formation was 1.1 × 10−5 per recipient spore of C5. The variation in frequency of exconjugant formation was also reported earlier (33) using a different plasmid, pTO1, carrying the φC31 int and att functions, and Streptomyces spp. An additional factor which influences exconjugant formation is the existence of a restriction system other than a methyl-specific restriction system (8). From these studies, it is evident that the exconjugant frequency varies with the type of recipient strain used for intergeneric conjugation irrespective of the E. coli donor. Experiments using S. peucetius ATCC 29050 spores as the starting material for conjugation failed, even though mycelial fragments yielded exconjugants. The heat-sensitive nature of the germinating spores could be the reason for this failure, as has been demonstrated for Streptomyces virginiae ATCC 13161 spores (33).
The vectors carrying the φC31 int and att functions integrated at a unique site in the S. ambofaciens chromosome, unlike pSAM2-based vectors (12). In addition, plasmids and cosmids carrying the φC31 int and att functions were found to produce stable exconjugants which could be propagated without detectable loss of plasmid markers (16). Southern hybridization analysis indicated that the plasmid pSET152 integrated at a unique site in the genomes of S. peucetius and C5. This observation was confirmed by cloning and sequencing of one of the recombination sites. The integration site was cloned from three different exconjugants in the case of C5 and from two different exconjugants for S. peucetius. All of the C5 clones released a 2.3-kb fragment when digested with SphI. The two independent clones from S. peucetius released a 2.2-kb fragment when digested with SphI. Comparison of the sequence of the C5 attachment site with sequence data of other attachment sites revealed 89 and 88.5% identity with attR junction sequences of S. coelicolor and S. lividans, respectively. The hybrid attachment site from S. peucetius revealed 87.4% identity with the attR junction attachment sites of S. lividans and S. coelicolor. The C5 and S. peucetius attachment sequences showed 88 and 93% identity, respectively, with the gene that encodes a chromosomal condensation protein of S. coelicolor. Sequence analysis clearly indicates that the φC31 attachment sites from various Streptomyces species are highly conserved. The core trinucleotide, TTG, was shown to be conserved in all att sites of Streptomyces, but in S. peucetius and C5, the core trinucleotide sequence was found to be TTC.
Cosmid pOJ436 carrying a large (30-kb) insert could integrate in the S. spinosa genome, apparently by homologous recombination, even though the cosmid carried the φC31 att site and integration functions (16). These data imply that homologous recombination could occur in the presence of int and att sites. Further, this study reported that cosmid pOJ436 and pOJ436 containing a small (2-kb) insert of S. spinosa DNA integrated at low frequencies at the attB site. In order to discover the effect of the presence of homologous sequences in cis on site-specific integration of the plasmid pSET152, a 3.7-kb DNA fragment (dnrUVJI) from the S. peucetius anthracycline biosynthetic pathway cluster was introduced into pSET152, and the resulting plasmid was mobilized from E. coli ET12567(pUZ8002/pPD1) to S. peucetius and C5. Southern analysis of 10 randomly selected exconjugants showed that the integration occurred only at the attB site. The presence of homologous sequences in addition to the att-int region apparently is species specific, since pSET152 integrated at the attB site in S. lividans even if a 15-kb homologous sequence was present (data not shown). However, we cannot rule out the role of clone-specific variation in homology-driven integration. These results indicate that site-specific recombination is not affected by the presence of homologous DNA sequences in S. peucetius and Streptomyces sp. strain C5.
The establishment of a gene transfer system and the stable integration of cloned genes at specific loci in the genome of S. peucetius prompted us to introduce the chitinase gene, chiC, cloned from a chitinase-overproducing mutant of S. peucetius (SPVI), as well as wild-type chiC. We reasoned that the introduction of an additional copy of the chiC gene in the genome of S. peucetius would facilitate the efficient growth of the recombinant strain on medium containing chitin. The culture supernatant of the recombinant strain SPD2 carrying SPVI chiC showed a >30-fold increase in chitinase activity compared to that of the wild-type strain, whereas the exconjugant SPD1, which carries wild-type chiC as an extra copy, showed only a 3.8-fold increase in chitinase activity. The net increase in chitinase activity due to the introduction of an additional copy of the chiC gene is apparently more than the expected levels in SPD2, as well as SPD1. SDS-PAGE analysis also confirmed this result, since a protein with a molecular mass of 42 kDa was induced at high levels in SPD1 and SPD2 compared to S. peucetius, and in SPD1, the induction was less than in SPD2. In addition, polyclonal antibodies raised against chitinase purified from S. peucetius reacted against chitinase produced from S. peucetius, SPVI, SPD1, and SPD2. Integration of chiC containing pSET152 did not affect antibiotic production in exconjugants (unpublished results), indicating the usefulness of this approach in antibiotic production, as well as chitinase production, on a commercial scale.
Acknowledgments
S.P. acknowledges with thanks the Council of Scientific and Industrial Research, New Delhi, India, for the award of a Senior Research Fellowship. This research was supported by a Centre for Genetic Engineering and Strain Manipulation grant from the Department of Biotechnology, Government of India, New Delhi, India, to K. Dharmalingam (BT/03/002/87-Vol.III).
We thank W. Strohl and Iain S. Hunter for providing plasmids and bacterial strains. We thank M. Ram Mohan for DNA sequencing.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltz, R. H. 1978. Genetic recombination in Streptomyces fradiae by protoplast fusion and cell regeneration. J. Gen. Microbiol. 107:93-102. [DOI] [PubMed] [Google Scholar]
- 3.Baltz, R. H. 1997. Molecular genetic approaches to yield improvement in actinomycetes, p. 49-62. In W. R. Strohl (ed.), Biotechnology of antibiotics, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 4.Baltz, R. H. 1998. Genetic manipulation of antibiotic-producing Streptomyces. Trends Microbiol. 6:76-82. [DOI] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Dekleva, M. L., J. A. Titus, and W. R. Strohl. 1985. Nutrient effects on anthracycline production by Streptomyces peucetius in a defined medium. Can. J. Microbiol. 31:287-294. [DOI] [PubMed] [Google Scholar]
- 7.Dharmalingam, K. 1986. Experiments with M13: gene cloning and DNA sequencing. Macmillan India Ltd., Madras, India.
- 8.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 9.Fouces, R., M. Rodriguez, E. Mellado, B. Diez, and J. L. Barredo. 2000. Conjugation and transformation of Streptomyces species by tylosin resistance. FEMS Microbiol. Lett. 186:319-325. [DOI] [PubMed] [Google Scholar]
- 10.Giebelhaus, L. A., L. Frost, E. Lanka, E. P. Gormley, J. E. Davies, and B. Leskiw. 1996. The Tra2 core of the IncPα plasmid RP4 is required for intergeneric mating between Escherichia coli and Streptomyces lividans. J. Bacteriol. 178:6378-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, England.
- 12.Kushtoss, S., M. A. Richardson, and R. N. Rao. 1991. Plasmid cloning vectors that integrate site-specifically in Streptomyces spp. Gene 97:143-146. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Lee. Y.-H. W., Z.-Y. Tzecheng, S.-C. Wang, W.-L. Cheng, and C. W. Chen. 1986. Structural instability of heterologous genes cloned in Streptomyces plasmid pIJ702. Biochem. Biophys. Res. Commun. 140:372-378. [DOI] [PubMed] [Google Scholar]
- 15.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 16.Matsushima, P., M. C. Broughton, J. R. Turner, and R. H. Baltz. 1994. Conjugal transfer of cosmid DNA from Escherichia coli to Saccharopolyspora spinosa: effects of chromosomal insertions on macrolide A83543 production. Gene 146:39-45. [DOI] [PubMed] [Google Scholar]
- 17.Matsushima, P., and R. H. Baltz. 1996. A gene cloning system for ‘Streptomyces toyocaensis'. Microbiology 142:261-267. [DOI] [PubMed] [Google Scholar]
- 18.Mazodier, P., R. Petter, and C. J. Thompson. 1989. Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 171:3583-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyashita, K., T. Fuji, and Y. Sawada. 1991. Molecular cloning and characterization of chitinase genes from Streptomyces lividans 66. J. Gen. Microbiol. 137:2065-2072. [Google Scholar]
- 20.Molnár, I., K.-P. Choi, N. Hayashi, and Y. Murooka. 1991. Secretory overproduction of Streptomyces cholesterol oxidase by Streptomyces lividans by a multi-copy shuttle vector. J. Ferment. Bioeng. 72:368-372. [Google Scholar]
- 21.Motamedi, H., A. Shafiee, and S.-J. Cai. 1995. Integrative vectors for heterologous gene expression in Streptomyces spp. Gene 160:25-31. [DOI] [PubMed] [Google Scholar]
- 22.Oakley, B. R., D. R. Kirsh, and N. R. Morris. 1980. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal. Biochem. 105:361-363. [DOI] [PubMed] [Google Scholar]
- 23.Paget, M. S. B., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smokvina, T., P. Mazodier, F. Boccard, C. J. Thompson, and M. Guerineau. 1990. Construction of a series of pSAM2-based integrative vectors for use in actinomycetes. Gene 94:53-59. [DOI] [PubMed] [Google Scholar]
- 27.Strauss, D. G. 1970. Anthracyclines—modern tumour-inhibiting agents. Folia Microbiol. (Prague) 23:152-161. [DOI] [PubMed] [Google Scholar]
- 28.Sun, Y., X. Zhou, J. Liu, K. Bao, G. Zhang, G. Tu, T. Kieser, and Z. Deng. 2002. ′Streptomyces nanchangensis', a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 148:361-371. [DOI] [PubMed] [Google Scholar]
- 29.VanWezel, G. P., J. White, G. Hoogvliet, and M. J. Bibb. 2000. Application of redD, the transcriptional activator gene of the undecylprodigiosin biosynthetic pathway, as a reporter for transcriptional activity in Streptomyces coelicolor A3(2) and Streptomyces lividans. J. Mol. Microbiol. Biotechnol. 2:551-556. [PubMed] [Google Scholar]
- 30.Vetrivel, K. S., and K. Dharmalingam. 2000. Isolation of a chitinase overproducing mutant of Streptomyces peucetius defective in daunorubicin biosynthesis. Can. J. Microbiol. 46:956-960. [PubMed] [Google Scholar]
- 31.Vetrivel, K. S., S. K. Pandian, U. Chaudhary, and K. Dharmalingam. 2001. Purification, cloning, and DNA sequence analysis of a chitinase from an overproducing mutant of Streptomyces peucetius defective in daunorubicin biosynthesis. Can. J. Microbiol. 47:179-187. [PubMed] [Google Scholar]
- 32.Voeikova, T. A. 1999. The conjugal transfer of plasmids from Escherichia coli to various strains of the order Actinomycetales. Russ. J. Genet. 35:1398-1404. [PubMed] [Google Scholar]
- 33.Voeykova, T., L. Emelyanova, V. Tabakov, and N. Mkrtumyan. 1998. Transfer of plasmid pTO1 from Escherichia coli to various representatives of the order Actinomycetales by intergeneric conjugation. FEMS Microbiol. Lett. 162:47-52. [DOI] [PubMed] [Google Scholar]
- 34.Volchegursky, Y., Z. Hu, L. Katz, and R. McDaniel. 2000. Biosynthesis of the antiparasitic agent megalomicin: transformation of erythromycin to megalomicin in Saccharopolyspora erythraea. Mol. Microbiol. 37:752-762. [DOI] [PubMed] [Google Scholar]