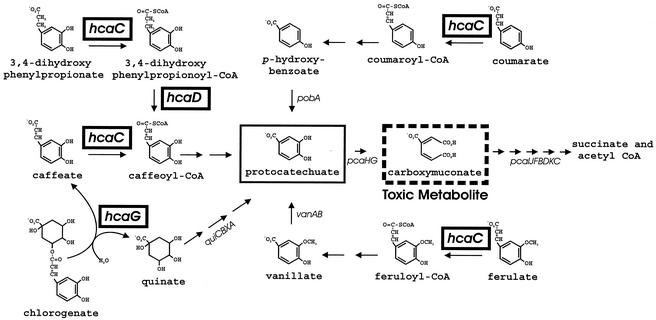
FIG. 1.
Metabolic pathways converging upon protocatechuate in Acinetobacter sp. strain ADP1. The ΔpcaBDK1 deletion blocks metabolism of carboxymuconate (dashed box). Accumulation of this compound prevents growth in the presence of protocatechuate (box) or its metabolic precursors, and this allows selection of strains with secondary mutations that prevent metabolism of these compounds (20). Replacement of ΔpcaBDK1 with wild-type DNA produces strains in which the secondary mutation is the only genetic barrier to catabolism. In previous investigations researchers reported properties of mutants blocked in expression of pcaHG (10, 20), pobA (14, 26), and vanAB (43, 56). In this report we describe a spontaneous hcaC (box) mutant unable to metabolize caffeate. As shown here, the ΔhcaC1 mutation appears to inactivate a CoA ligase required for growth with coumarate, ferulate, and 3,4-dihydroxyphenylpropionate, as well as caffeate. Between hcaC and pobA in the Acinetobacter chromosome are the structural genes hcaD (box), encoding an enzyme that oxidizes 3,4-dihydroxyphenylpropionyl-CoA to caffeoyl-CoA, and hcaG (box), encoding an enzyme that hydrolyzes chlorogenate to quinate and caffeate. Wild-type Acinetobacter cells exhibit doubling times ranging from 40 min to 1 h when they are grown at the expense of 3,4-dihydroxyphenylpropionate, coumarate, ferulate, quinate, caffeate, or chlorogenate.