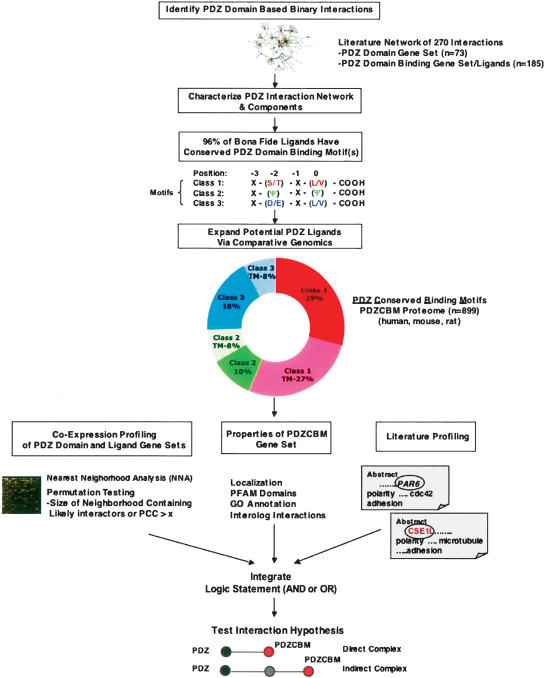
Figure 1.
Computational schema to identify PDZ ligands and PDZ complexes. Step (1): Created database of 270 binary PDZ domain-based ligand interactions from manual literature curation. Step (2): Analyzed characteristics of PDZ–ligand interaction network including expression correlation between PDZ-encoding gene and ligand gene sets, as well as properties of bona fide ligands (e.g., level of PDZ-binding motif conservation). Step (3): Identified that 96% of literature-identified ligands have conserved PDZ-binding motifs among three mammalian genomes (human, rat, mouse). Of these, 79% match one of three “canonical” consensus carboxy-terminal motifs. The canonical consensus motifs shown for each of the three classes were derived from reviews (Sheng and Sala 2001; Nourry et al. 2003). Step (4): Performed systematic comparative genomic analysis of three mammalian proteomes to reveal an 899-member gene set that encodes proteins with canonical PDZ Conserved Binding Motifs (PDZCBM). The motif class distribution and percentage of each class predicted to have a transmembrane (TM) domain by the TMHMM algorithm is shown (Krogh et al. 2001). Step (5): Pairwise relationships among PDZ domain-encoding genes and the PDZCBM gene set are determined from correlated mRNA levels, cellular localization, and common literature co-citation patterns between a PDZ gene and potential ligand. Criteria for significant co-expression levels are based on nearest neighborhood analysis (NNA) indexes and/or a Pearson’s correlation coefficient (PCC) threshold. Orthologous information from model organisms (e.g., interologs) is mined to derive potential PDZ–ligand interactions. Step (6): All data are integrated from these diverse data types employing logical operators AND or OR to provide testable hypothesis as to PDZ complexes. Step (7): The predicted interactions are tested in mammalian cells by co-immunoprecipitation, analyzing the effects of mutating interaction motifs/domains, and/or functional assays.