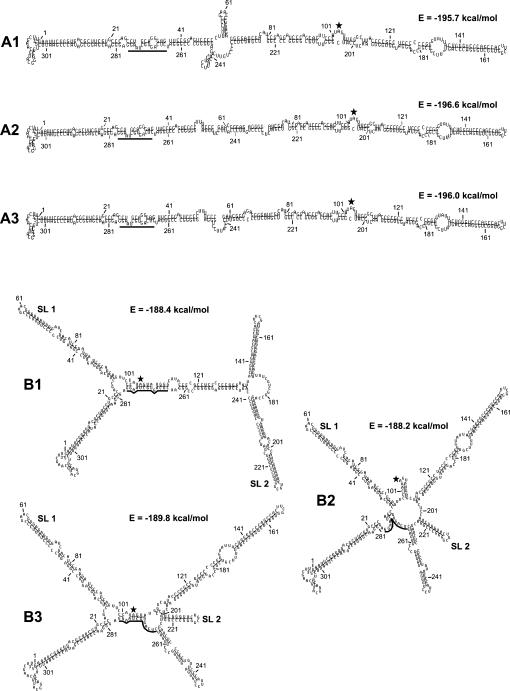
FIGURE 2.
Drawings of the unbranched (A1–A3) and branched (B1–B3) secondary structures predicted to be formed by MD-III-2 RNA. The structures and energies shown were calculated using the massively parallel genetic algorithm MPGAfold (Shapiro and Navetta 1994; Shapiro and Wu 1996, 1997; Wu and Shapiro 1999; Shapiro et al. 2001a,b). Represented here are the dominant structures from the final solutions of MPGAfold runs in all population levels (see Table 1). Numbering refers to sequence positions in the miniaturized cDNA clone pMD-III-2. The amber/W adenosine at position 104 is indicated by a black, five-pointed star. The black bar indicates sequence positions 266–278, which are base-paired around the amber/W site in branched structure B1. SL1 and SL2 indicate stem–loops previously found to be important for editing (Casey 2002). First positions in terminal loops are as follows: 59 in SL1, and 212 in SL2.