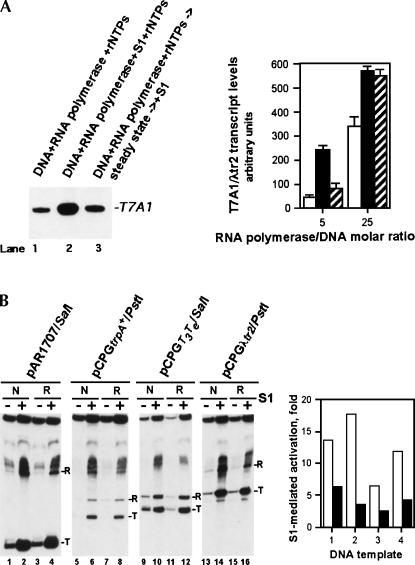
FIGURE 3.
Effect of delayed addition of S1 to in vitro transcription reactions and the effect of S1 on the efficiency of transcription termination. (A) The delayed addition of S1 to in vitro transcription reactions results in greatly reduced transcription-stimulatory activity. The quantitated results of two independent, parallel experiments are shown. Reactions in lanes 1 (open columns) and 2 (black columns) are similar to those in lanes 5 and 10 of Figure 1A; in lane 3 (hatched columns), purified native S1 was added 30 min after the initiation of the in vitro transcription reaction, and the reactions were incubated at 37°C for an additional 60 min. The reactions were performed with an approximately fivefold (see Materials and Methods) or 25-fold molar excess of RNA polymerase over the supercoiled pCPGλtr2 DNA template. The RNA polymerase/DNA template molar ratios are indicated. Incubation of RNA polymerase alone at 37°C up to 90 min produced no significant reduction in the enzyme's activity. (B) Mild stimulation of the transcription termination efficiency in the presence of 0.3 μM purified native S1 (lanes “N”) or 0.6 μM recombinant His-tagged S1 (lanes “R”). RNA transcripts terminated at specific terminators (T) and runoff RNA transcripts (R) are indicated. The quantitated results of the experiment are shown on the right; open and black columns show the magnitudes of the S1-mediated transcriptional activation for RNA transcripts terminated at specific terminators and runoff RNA transcripts, respectively. The linearized plasmid DNA templates used in this experiment were pAR1707 (1), pCPGtrpA + (2), pCPGT3Te (3), and pCPGλtr2 (4). DNA templates carried the T7A1 promoter followed by a defined transcription terminator downstream (the type of terminator is apparent from the plasmids' names); all fourtemplates are described in Reynolds et al. (1992).