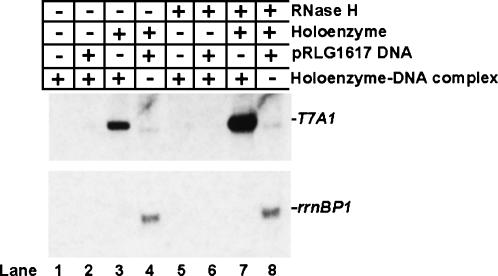
FIGURE 4.
Inactivation of RNA polymerase rather than the DNA template is the principal reason for poor transcriptional cycling in in vitro transcription reactions with supercoiled DNA. Five microliters of 500 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 1 M NaCl were mixed with 25 μL of purified water, 5 μL RNA polymerase holoenzyme (1.2mg/mL), 5 μL pCPGλtr2 supercoiled DNA (0.89 mg/mL) with the T7A1 promoter and λtr2 terminator, 10 μL of 5× rNTP mix containing 1 mM each of ATP, GTP, CTP, and UTP, and 0.01 μCi of [α32P] UTP. Following a 30-min incubation at 37°C, the entire 50-μL reaction mixture was passed through a Superdex 200 HR 10/30 column pre-equilibrated with TGED buffer containing 100 mM NaCl; 0.5-mL fractions were collected. Aliquots of 250 μL from fractions containing the DNA-bound transcription complexes were concentrated to ∼25 μL using Microcon-10 concentrators (Amicon), and 3-μL aliquots of the resulting purified RNA polymerase–DNA complex were used in in vitro transcription reactions, performed as described in Materials and Methods, with the exception that RNA polymerase and the DNA template were omitted. When indicated, RNA polymerase holoenzyme was added to a final concentration of 0.05 mg/mL; pRLG1617 supercoiled DNA template with the ribosomal rrnBP1 promoter and T1T2 terminator (Ross et al. 1990) to 0.08 mg/mL; and RNase H (New England Biolabs), 5 U per 20 μL reaction. A representative result of two independent experiments is shown.