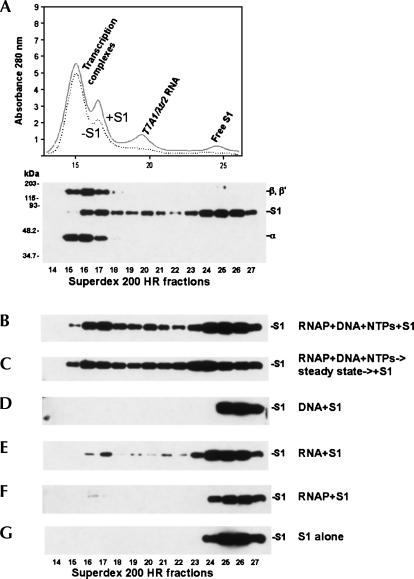
FIGURE 5.
S1 interacts with transcription complexes. (A) Top: the A280 profile resulting from size-separation of an in vitro transcription reaction mixture on a Superdex 200 HR 10/30 column. The supercoiled pCPGλtr2 DNA template (Reynolds et al. 1992) used in this set of experiments carried the T7A1 promoter and λtr2 terminator. The individual components of the reaction were identified as described in Materials and Methods. Bottom: S1 and RNA polymerase subunits inthe fractions were also identified by Western blot analysis using S1-specific and core RNA polymerase-specific antibodies. (B,C) The in vitro transcription reactions were similar to those described in Materials and Methods, with the exceptions that 0.24× (12 μL) reactions were utilized here, and the reactions contained 0.2 μM purified native S1. The reaction ingredients are indicated at the right. S1 in the Superdex 200 HR 10/30 column fractions is visualized here and below by immunoblotting with S1-specific antibodies. (D) 0.2 μM S1 plus pCPGλtr2 supercoiled DNA (0.089 mg/mL). (E) In vitro transcription reactions similar to those described above were carried out and, after a 30-min incubation at 37°C, 4 U of RNase-free DNase (Promega) was added to the mixtures to digest the DNA template. Following another 30-min incubation at 37°C, the reactions were deproteinized by phenol extraction, and the aqueous phase was precipitated with ethanol. The RNA pellet was redissolved in 12 μL of 1× TB containing 0.2 μM purified native S1, and the binding experiments were performed as described in Materials and Methods. The substitution of DNase with a DNase/RNaseH mixture in the above purification procedure produced similar S1–RNA binding patterns, suggesting that DNA–RNA hybrids are unlikely to play a major role in the interaction of S1 with transcription complexes (data not shown). (F) 0.2 μM S1 plus 1.3 μM RNAP holoenzyme. In a similar binding assay, Δ335–556 S1 showed no detectable complex formation with the polymerase. (G) 0.24 μM S1.