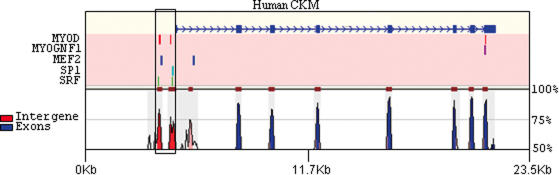
Figure 3.
Predicted TF-binding sites in human–mouse conserved regions around the CKM (creatine kinase, muscle) gene. The genomic regions, along with 5 kb upstream and 2 kb downstream, of the CKM gene were extracted from the human and mouse genomes and aligned using the BLASTZ software (151). The BLASTZ alignments were then fed into the rVISTA program (153) through the website http://rvista.dcode.org. Binding sites for several TFs that are known to regulate gene expression in the muscle tissue were then predicted on the human sequence using the PWM models available from the TRANSFAC database (12). The predicted sites can be dynamically viewed and clustered through the above website. For the purpose of this current figure, we required that at least two binding sites belonging to different TFs be present within a window of 100 nt. A cluster of sites was observed in the immediate 5′ upstream region of this gene (boxed). Percent conservation between the two sequences is shown; regions with ≥75% conservation are colored. The human gene structure is shown at the top in blue.