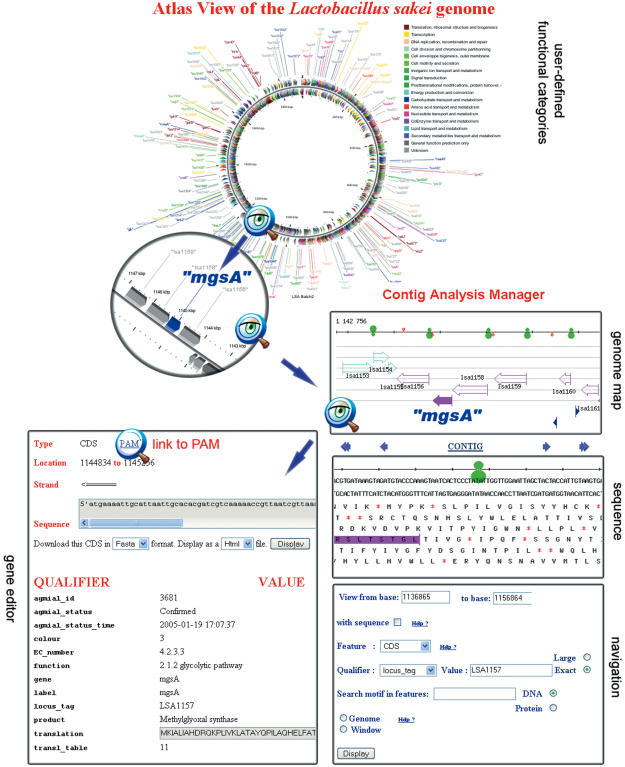
Figure 1.
Views of the DNA sequence at different scales. The upper part of the figure represents an atlas view of the genome obtained with CGView. One can zoom on a particular region of this map, for instance on the area containing the mgsA gene: a methylglyoxal synthase that belongs to the glycolytic pathway. Clicking on this gene will open the MuGeN interface showing its genomic context (genome map frame). It is possible to zoom on this representation to see the DNA sequence and the translation in the six reading frames (sequence frame). The green symbol represents the RBS, the gene sequence is colored as in the previous view. The navigation window allows one to move along the genome, either by entering a range of base numbers, or by looking for a feature with a particular qualifier or by specifying a DNA or protein motif to be searched for in the current window or in the complete genome. The window at the lower left of the figure shows the gene editor. Most fields are automatically filled, in particular the gene annotation qualifiers since, in general, CDS annotation is performed in PAM and then updated in CAM (see Figure 4). Clicking on the link to PAM, indicated by the magnifying glass, will lead the annotator to the PAM interface shown in Figure 2. For clarity the Artemis interface is not shown on the figure.