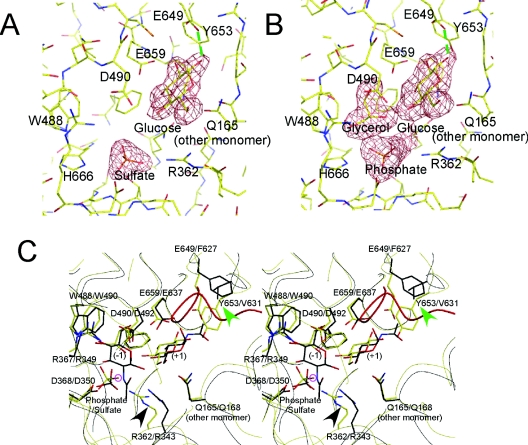
Figure 2. Active site structure of CgCBP.
(A) Wireframe model of CgCBP–SO4, and the unbiased |Fobs| − |Fcalc| electron-density map of the bound glucose and phosphate (3.0σ). The residues (single-letter amino acid codes are used) involved in subsite formation (R362, W488, E649, Y653, E659, and Q165 from the adjacent subunit), sulphate recognition (H666), and catalysis (D490) are labelled. Glucose takes on the β-anomer configuration and forms a hydrogen bond with E649 (green broken line). (B) Wireframe model of CgCBP–PO4, and the unbiased |Fobs| − |Fcalc| electron-density map of the bound glucose, glycerol, and phosphate (3.0σ). (C) Stereo view of CgCBP–PO4 and VpChBP superimposed at the active site. The backbone traces of CgCBP and VpChBP are coloured yellow and black, respectively. The bound glucose, glycerol, and phosphate ion in CgCBP (yellow), and the GlcNAc molecules and sulphate ion in VpChBP (black) are shown as wireframe models. Subsites −1 and +1 are labelled (−1) and (+1), respectively. The structurally conserved residues are shown as a wireframe model and labelled in the order corresponding to CgCBP and VpChBP. The 495–513 loop in CgCBP is coloured red. The residues related to N-acetyl/hydroxy group recognition at the C-2 position in each subsite are indicated by arrowheads. The oxygen atom suitable for direct attack on the C-1 atom of glucose at subsite −1 is circled in pink. Single-letter amino acid codes are used.