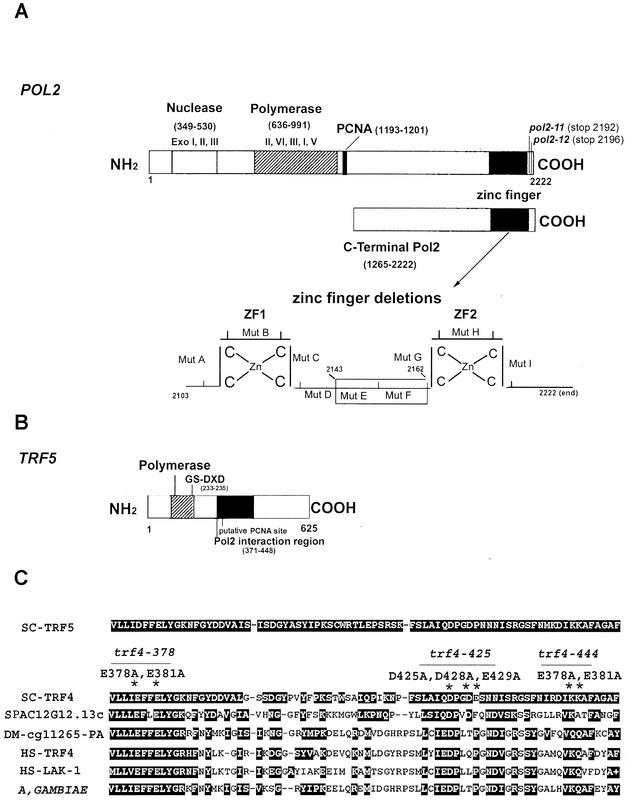
FIG. 1.
(A) Schematic diagram of the POL2 gene describing conserved exonuclease domains and polymerase domains. The C-terminal portion of Pol2 used for the two-hybrid library screen includes aa 1265 to 2222. Ten amino acid deletions in the zinc finger region of Pol2 were used to map the interaction of Trf5; the zinc finger deletion region is shown enlarged. (B) Schematic diagram of the TRF5 gene. The conserved nucleotidyl transferase domain occurs between aa 178 and 241. The principle conserved motif is from aa 217 to 235. Aspartates 233 and 235 in Trf5 correspond to aspartates 236 and 238 in the DXD motif in Trf4, and since mutations in these residues reduce the DNA polymerase activity of Trf4, we have labeled this the putative active site (6, 70). (C) The 78-aa POL2 interaction region of TRF5 homologous to TRF genes in yeast, human, and fly. The alignment was made using BLAST data from the SGD and GenBank. SC, S. cerevisiae; SP, Schizosaccharomyces pombe; DM, Drosophila melanogaster; HS, Homo sapiens. The corresponding amino acid numbers and accession numbers are SC-Trf5_1050861 (aa 371 to 448); SC-Trf4_950226 (aa 374 to 451); SP-C12G12.13C_2130260 (aa 1006 to 1081); DM-CG11265_22831959 (aa 461 to 538); HS-Trf4_5565687 (aa 105 to 182); HS-Lak-1_5139669 (aa 181 to 258); A. gambiae_21288943 (aa 460 to 536). Both identities and similarities are highlighted. The asterisks identify mutations in trf4 that cause lethality (trf4-378; trf4-425; trf4-444).