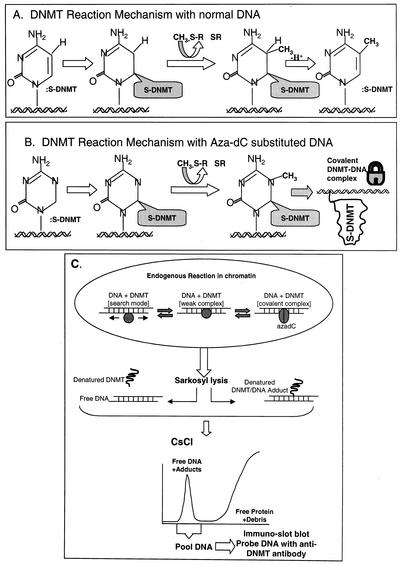
FIG. 1.
DNMT reaction mechanism and the ICM assay. (A) Methylation on non-aza-dC-containing DNA proceeds via a nucleophilic attack by cysteine thiolate of DNMT at the C-6 on cytosine followed by a second nucleophilic attack at C-5 by the methyl group of S-adenosylmethionine (methyl donor). This results in the transfer of the methyl group to C-5 and an intermediate that is resolved by β elimination of DNMT at C-6 and abstraction of a proton from C-5. (B) Mechanism of action on aza-dC-substituted DNA involves a methyl transfer at the N-5 of aza-dC and formation of a stable or covalent complex between enzyme and DNA that is resistant to ionic detergents, high ionic strength, and temperature (viz., conditions that dissociate proteins that are noncovalently bound to DNA) (9, 10, 15, 34). (C) Overview of the ICM assay. The endogenous DNMT reaction in vivo shows the enzyme in either a one-dimensional search mode (scanning for suitable CpG targets in chromatin) in a weak binding mode (enzyme paused over a chromatin-accessible CpG site in vivo) or in the covalent DNMT-DNA complex mode with aza-dC-substituted DNA. Direct addition of Sarkosyl lyses the cells and disrupts weak (noncovalent) protein-DNA complexes in chromatin; however, aza-dC-induced DNMT-DNA complexes are not dissociable under these conditions. The viscous lysate is sheared and overlaid onto a step CsCl gradient (Materials and Methods) that resolves DNA from the bulk, excess free protein and debris in a single step (35, 38, 39). Proteins stably bound to the DNA are dragged down to the 1.7-g/ml fraction of the gradient. The DNA fraction contains DNMTs only when the cells have been prelabeled with aza-dC. Specific DNMTs are quantified in the DNA peak by Western slot blotting. Note that the material at the top of the gradient gives a nonspecific Western blot signal due to the excess of membranous debris and cannot be relied on to measure free DNMT. To demonstrate that the DNA peak-associated DNMT signal is real (and not carryover from the top of the gradient), the DNA peak can be collected and rebanded in CsCl to give quantitative recovery of the DNMT signal in a second gradient (data not shown). The ICM data were not affected by digestion of lysates with RNase A prior to centrifugation; therefore, RNase digestion was deemed unnecessary.