Fig. 3.
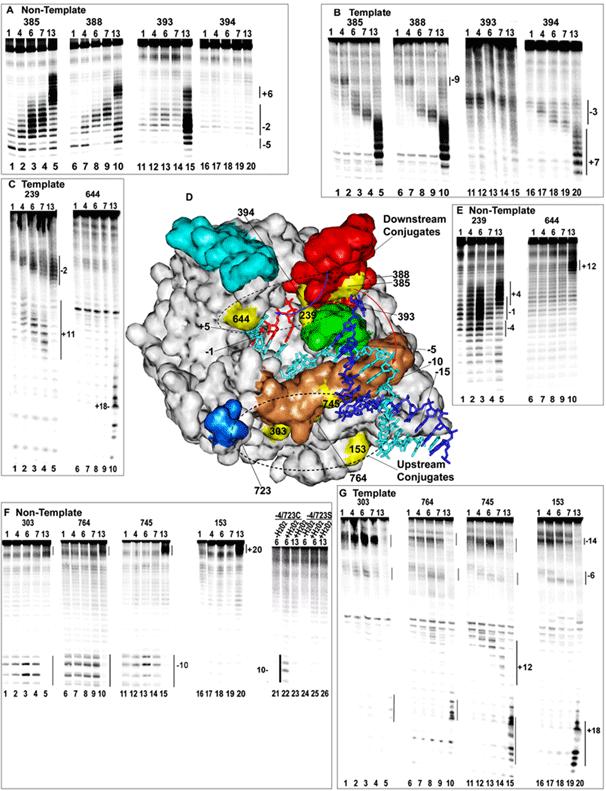
DNA cleavage by Fe-BABE tethered to introduced cysteines. A: Cleavage of transcription complexes with 5'-end labeled NT strand by conjugates at aa positions 385 (lanes 1-5), 388 (lanes 6-10), 393 (lanes 11-15), or 394 (lanes 16-20). The RNA length in the complex is 1, 4, 6, 7, or 13 nt as indicated at the top of each lane (a length of 1 corresponds to adding only 3'-dGTP). B: As in A but with 5'-labeled T strand. C: As in B, but with conjugates at either 239 (lanes 1-5) or 644 (lanes 1-10) D: Crystal structure of a T7RNAP ITC with a 3 nt RNA (pdb # 1QLN). NT strand is dark blue; T strand is cyan; RNA is red; the promoter recognition loop is tan; the thumb subdomain (aa 330-410) is red; the intercalating hairpin (aa 232-242) is green; the fingers 'flap' (aa 586-620) is cyan; and the 'positive patch' (K711/713/714) is blue. Positions of Fe-BABE conjugation are yellow and are labeled (aa 723 is hidden; 764 and 393 are largely obscured). Numbering of the DNA is relative to the +1 start site, and of the RNA is relative to the RNA 3'-nt. E: As in C, but with 5'-labeled NT strand. F: As in A but with conjugates at 303 (lanes 1-5), 764 (lanes 6-10), 745 (lanes 11-15), and 153 (lanes 16-2-0). Also shown is cleavage by an Fe-BABE conjugated enzyme from which 4 cysteines have been removed, but which retains a cys at 723 (lanes 21-23), and an enzyme in which C723 was mutated to ser (lanes 24-26). H2O2 was added to all reactions except those in lanes 21 and 24.. G: As in F but with 5'-labeled T strand.
