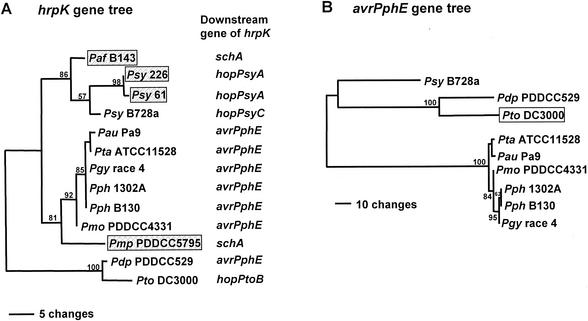
FIG. 6.
Phylograms of optimal trees of EELs in P. syringae strains based on 250 bp of hrpK (A) and full-length avrPphE (B) sequences. Horizontal branch length is proportional to the estimated number of nucleotide substitutions, and bootstrap probabilities (as percentages) are indicated above or below the internal branches. The trees are rooted at the midpoint. Abbreviations for strains used in the figure follow: Psy, pathovar syringae; Pdp, pathovar delphinii; Pto, pathovar tomato; Pau, pathovar angulata; Pmo, pathovar mori; Pph, pathovar phaseolicola; Pgy, pathovar glycinea. In panel A, strains with EELs encoding ShcA-HopPsyA homologs are shown in shaded boxes. In panel B, the open box indicates that the avrPphE homolog used in phylogenetic analysis is not associated with the EEL.