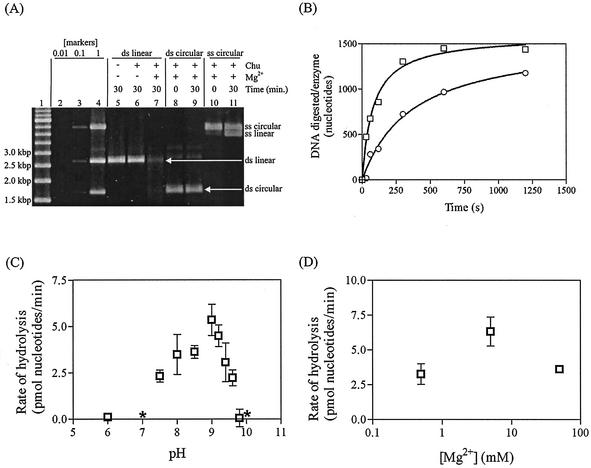
FIG. 5.
SPP1 Chu enzyme characterization. (A) DNA (2.3 nM ends or 1.15 nM molecules) was digested with refolded Chu (at least 2.3 nM, as determined by titration) for 30 min at 37°C under standard reaction conditions (see Materials and Methods). The gel was scanned to determine DNA quantities with an AlphaImager 2000. Lanes 1 through 4, molecular weight markers. Lane 1, 500-bp ladder; lanes 2, 3, and 4, combination of the substrates used in the assays, i.e., linear dsDNA, circular dsDNA, and circular ssDNA, at initial concentrations of 0.01×, 0.1×, and 1×, respectively. Lane 5, control without Mg2+ or Chu. (B) Phosphorylated (squares) or dephosphorylated (circles) PstI-cut pUC19 (0.151 nM ends) was incubated with Chu (at least 0.151 nM, as determined by titration) for 2 min at 37°C, and reactions were then initiated with 5 mM Mg2+. DNA was digested under standard reaction conditions for 0, 0.5, 1, 2, 5, or 10 min. The 0-min sample was quenched with 20 mM EDTA prior to the addition of Mg2+. (C) PstI-cut pUC19 (2.3 nM ends) was digested with refolded Chu (at least 2.3 nM, as determined by titration) for 30 min at 37°C under standard conditions, except that pH was varied. (D) PstI-cut pUC19 (2.3 nM ends) was digested with refolded Chu (at least 2.3 nM, as determined by titration) for 30 min at 37°C under standard conditions, except that Mg2+ concentration was varied. *, the mean value was below zero, but the rate fell within the range of 0.