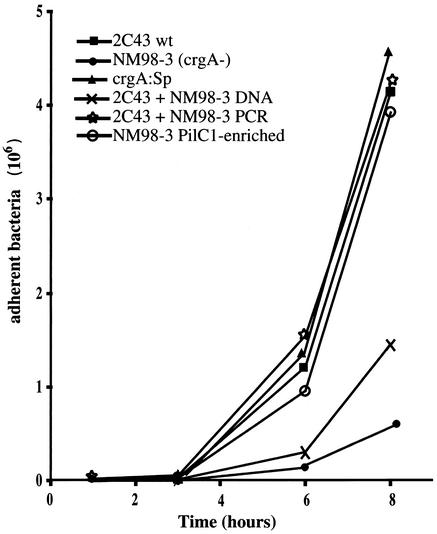
FIG. 4.
Adhesion of 2C43 and various crgA mutants on HUVECs. Cells were infected for 30 min with 105 bacteria in exponential growth phase and then washed. The monolayer was washed every hour. At different time points, monolayers were harvested and the number of adhesive bacteria was determined. 2C43 is the wild-type bacterium, NM98-3 is the previously described crgA mutant of 2C43 (4), crgA:Sp is the crgA mutant that we engineered in 2C43, 2C43+NM98-3 DNA corresponds to the strain 2C43 transformed with the total chromosomal DNA of NM98-3, 2C43+NM98-3 PCR corresponds to a transformant of strain 2C43 with the PCR product of the crgA mutation of NM98-3, and NM98-3 PilC1-enriched corresponds to cell-associated CFU of NM98-3 after 3 h of infection.