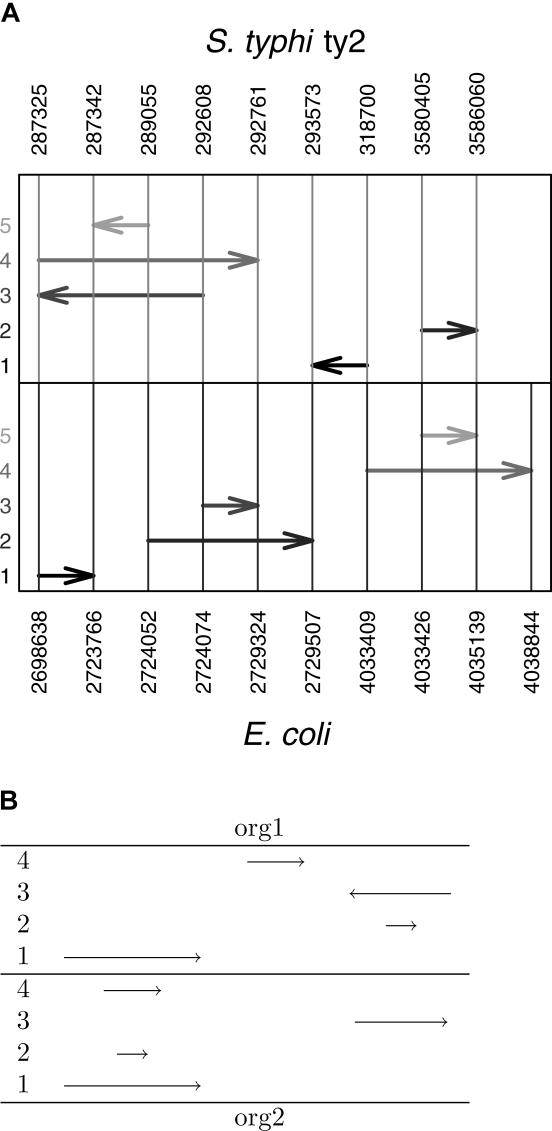
Figure 2. Schematic Examples of Situations That May Be Encountered While Clustering Entries of the Comprehensive Table (Steps 2 and 3 in the Mapping Phase).
An entry is represented as two arrows having the same label, one in the top and one in the bottom panels. Labels for the arrows are given on the left side. The relative direction of identically labeled arrows represents the relative orientation of the corresponding segments.
(A) A real example from the comparison of E. coli versus S. typhi ty2 demonstrating the difficulty of identifying the consecutive entry. In the top (bottom) panels, the positions of the segments in S. typhi ty2 (E. coli) are given. Arrows having the same label are drawn with identical grayscale color. The significantly overlapping entries, e.g., 2 and 3 in E. coli, correspond to rDNA operons, of which there are seven units in each organism. Each of these seven units have a hit overlapping, for example Entry 2 in both top and bottom panels. For the sake of clarity, the figure demonstrates only a part of the hits and is schematic (not to scale).
(B) Illustration of nuisance cross-overlaps and inparalogs. Entry 2 overlaps with Entry 1 in org1 and with Entry 3 in org2. Assuming that both overlaps are long enough and that Entry 2 is significantly shorter than either Entry 1 or Entry 3, Entry 2 is considered as a nuisance cross-overlap. On the other hand, Entry 4 overlaps Entry 1 in org2, but does not overlap with other entries in org1, and hence is not a nuisance cross-overlap. Assuming the length of Entry 1 is significantly greater than that of Entry 4, Entry 1 is considered to be the positional ortholog, while Entry 4 is considered to be the inparalog.