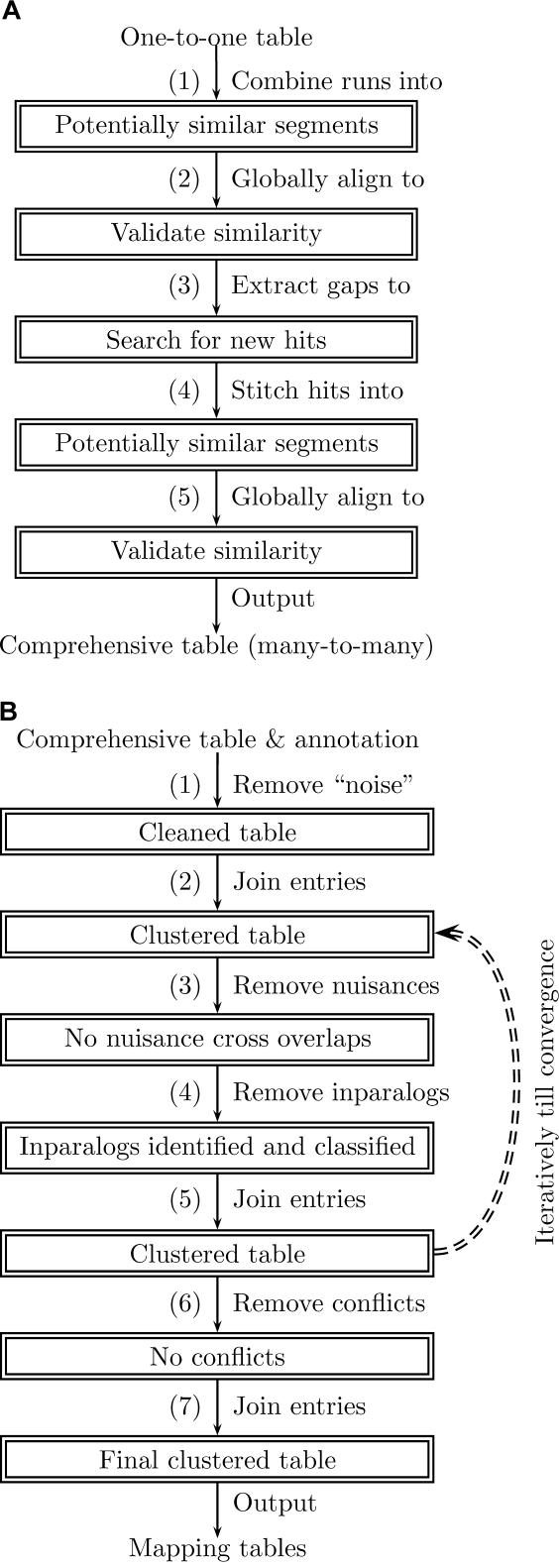
Figure 3. A Flow Diagram of MAGIC.
(A) A flow diagram of the preprocessing phase (see Preprocessing Phase: Building a Comprehensive Table of Similar Segments).
(A1) A one-to-one table (anchors) between two genomes is used to calculate runs of anchors corresponding to potentially similar segments.
(A2) These segments are globally aligned to validate their similarity and to find unmatched regions (these regions are candidates for indels or reordering events).
(A3) The unmatched regions are extracted (the entries remaining in the table correspond thus to maximal similar segments) and aligned (including uncovered regions between the runs) locally against the other genome to search for new hits, in an attempt to make the table comprehensive.
(A4) Local hits are stitched together to form potentially similar segments.
(A5) These segments are globally aligned to validate their similarity and to extract unmatched regions out of them. The resulting table is comprehensive and consists of maximal similar segments.
(B) A flow diagram of the mapping phase (see Mapping Phase: Clustering into RF Segments).
(B1) Short entries and entries corresponding to known selfish DNA in either of the genomes are removed.
(B2) Consecutive entries are joined for the first time (which makes it easier to identify nuisance cross-overlaps and inparalogs).
(B3) Nuisance cross-overlaps (see Steps 3–5: Identifying nuisance, classifying inparalogs, and re-clustering and Figure 2) are identified and discarded.
(B4) Inparalogs are identified, classified, and removed from the table.
(B5) Consecutive entries are joined again (nuisances and inparalogs may have hindered joining some of the entries in the previous clustering). Steps B3–B5 are performed iteratively until the table converges (no more nuisance or inparalogs are identified).
(B6) Remaining conflicts (unresolved significant overlaps), corresponding most likely to selfish DNA, are removed.
(B7) Consecutive entries are joined for the final time (the conflicts removed in B6 may have hindered joining some of the entries in the previous clustering). The output of the mapping phase consists of several tables, among which are: a one-to-one table describing the RF segments, a one-to-one table describing the positional orthologous segments contained in the RF segments, a table of the classified inparalogs in each organism, and a table of identified transposable elements.