Abstract
DNA mismatches are generated when heteroduplexes formed during recombination involve DNA strands that are not completely complementary. We used tetrad analysis in Saccharomyces cerevisiae to examine the meiotic repair of a base–base mismatch and a four-base loop in a wild-type strain and in strains with mutations in genes implicated in DNA mismatch repair. Efficient repair of the base–base mismatch required Msh2p, Msh6p, Mlh1p, and Pms1p, but not Msh3p, Msh4p, Msh5p, Mlh2p, Mlh3p, Exo1p, Rad1p, Rad27p, or the DNA proofreading exonuclease of DNA polymerase δ. Efficient repair of the four-base loop required Msh2p, Msh3p, Mlh1p, and Pms1p, but not Msh4p, Msh5p, Msh6p, Mlh2p, Mlh3p, Exo1p, Rad1p, Rad27p, or the proofreading exonuclease of DNA polymerase δ. We find evidence that a novel Mlh1p-independent complex competes with an Mlhp-dependent complex for the repair of a four-base loop; repair of the four-base loop was affected by loss of the Mlh3p, and the repair defect of the mlh1 and pms1 strains was significantly smaller than that observed in the msh2 strain. We also found that the frequency and position of local double-strand DNA breaks affect the ratio of mismatch repair events that lead to gene conversion vs. restoration of Mendelian segregation.
DNA mismatch repair (MMR) is the process by which base–base mismatches and certain other types of DNA lesions are corrected (reviewed by Harfe and Jinks-Robertson 2000). DNA mismatches can be formed either by errors made during DNA replication (polymerase slippage or misincorporation of bases by DNA polymerase) or by formation of recombination-associated heteroduplexes in which the DNA strands are not perfectly complementary. Correction of replication errors is sometimes termed the “spellchecker” function of MMR. The related process of correction of DNA mismatches within heteroduplexes results in the nonreciprocal recombination process of gene conversion. The mismatch repair proteins also have several roles that are not directly related to correction of mismatches (reviewed by Harfe and Jinks-Robertson 2000). First, a number of the mismatch repair proteins are involved in reducing the rate of recombination between diverged repeated sequences. Second, some MMR proteins act as DNA damage sensors in DNA damage checkpoint pathways (Stojic et al. 2004). Third, some MMR proteins promote crossing over (Börner et al. 2004; Hoffmann and Borts 2004). Finally, some MMR proteins participate in the removal of single-strand “tails,” allowing recombination between repeated genes by the single-strand annealing pathway (Sugawara et al. 1997). Our study is focused on the role of the MMR proteins in DNA mismatch repair.
In vivo and in vitro studies of mismatch repair in Escherichia coli (reviewed by Modrich and Lahue 1996) have defined the steps in mismatch repair and the proteins involved in catalyzing these steps. These steps include (1) the recognition of the DNA mismatch, (2) the identification of the newly synthesized DNA strand, (3) the nicking of the newly synthesized strand near the mismatch, (4) the excision of the mispaired DNA on the nicked strand, and (5) DNA synthesis and ligation to fill in the gap on the nicked strand. To initiate the process of repair, MutS binds to the DNA mismatch, and MutL facilitates a MutS/MutH interaction. This MutS/MutH complex stimulates MutH-mediated nicking of the nonmethylated, newly synthesized DNA strand. The nicked strand then is removed via helicase and exonuclease activities. DNA synthesis across the excised region and subsequent DNA strand ligation complete the repair process.
The process of mismatch repair is much more complicated in eukaryotes. Our understanding of the steps in the process and the proteins involved for each step is based on genetic studies (types of mutations observed in strains lacking MMR and the efficiency of correction of various types of mismatches formed during recombination) and biochemical studies. All steps in the process have not yet been defined. In Saccharomyces cerevisiae, six MutS homologs (Msh1–6p), four MutL homologs (Mlh1–3p, Pms1p), and no MutH homologs have been identified (reviewed by Harfe and Jinks-Robertson 2000). Most eukaryotic MMR and related processes are carried out by different combinations of a heterodimer of MutS homologs and a heterodimer of MutL homologs. For instance, base–base mismatches as well as one-base insertions or deletions are repaired by an Msh2p/Msh6p/Mlh1p/Pms1p complex, whereas small (1–14 bases) loops are repaired by an Msh2p/Msh3p/Mlh1p/Pms1p complex. Heterodimers of Mlh1p and Mlh2p or Mlh1p and Mlh3p play a minor role in the repair of frameshift mutations generated as a result of polymerase slippage at mononucleotide runs (Harfe et al. 2000). In contrast, Msh4p and Msh5p do not have any known spellchecker activity, but these proteins (as well as Mlh1p and Mlh3p) are involved in promoting crossovers (Hunter and Borts 1997; Wang et al. 1999; Börner et al. 2004). Finally, Msh1p functions exclusively in the mitochondria (Reenan and Kolodner 1992; Sia and Kirkpatrick 2005).
While mismatch recognition has been well characterized, less is understood regarding the strand discrimination signal and subsequent processing steps of MMR in eukaryotes. It has been proposed that proliferating cell nuclear antigen (PCNA), the sliding clamp associated with DNA polymerase, may differentiate which strand is the template for repair (Umar et al. 1996). In an in vitro system of MMR, the 3′–5′ excision reaction requires PCNA and the replication factor C clamp loader (Dzantiev et al. 2004). It is also possible that single-strand nicks left during Okazaki fragment formation might direct repair (Genschel et al. 2002).
A number of nucleases have been implicated as involved in MMR: Rad1p/Rad10p (the yeast equivalent of XPF/ERCC1), Exo1p, Rad27p (the yeast homolog of the Fen1 flap endonuclease), and the proofreading activity of DNA polymerase δ (reviewed by Marti and Fleck 2004). Of these nucleases, Exo1p is the one most likely to have a direct role in MMR, since mutations in EXO1 result in a mutator phenotype (Tishkoff et al. 1997). Since this mutator phenotype is much weaker than that observed in msh2, mlh1, or pms1 strains, and since the types of mutations observed in exo1 strains are substantially different from those observed in strains with other MMR mutations, the Exo1p activity is likely to be partly functionally redundant with that of one or more other nucleases (Tishkoff et al. 1997; Tran et al. 2001). It is also likely that at least part of the mutator phenotype of exo1 mutants is independent of an effect on MMR (Tran et al. 2001). In addition, Exo1p physically interacts with Msh2p, Msh3p, and Mlh1p (Tishkoff et al. 1997; Tran et al. 2001). Finally, in vitro studies indicate that Exo1p is required for bidirectional repair of a mismatch (Constantin et al. 2005).
Mismatches resulting from misincorporation errors of DNA polymerase are highly variable in type and position. In contrast, the mismatches in heteroduplexes formed during meiotic recombination are completely defined in type and position. In this study, we construct yeast strains that form either base–base (A/A or T/T) mismatches or four-base loops in heteroduplexes generated during meiotic recombination. By tetrad analysis (details explained in the results), we examined the efficiency of repair of these two types of mismatches in wild-type strains and in strains with MMR defects. We investigated the effects of all of the MutS and MutL homologs, except the mitochondria-specific MSH1 gene product. In addition, we analyzed the effects of mutations in the nuclease-encoding RAD1, EXO1, and RAD27 genes, and a mutation in the proofreading nuclease of DNA polymerase δ.
This study is the first in which the effects of all of the genes previously implicated in DNA mismatch repair are analyzed for specific types of DNA mismatches in one isogenic genetic background. Our results confirm the importance of the Msh2/Msh6/Mlh1/Pms1 proteins in the repair of base–base mismatches and the Msh2/Msh3/Mlh1/Pms1 proteins in the repair of a four-base loop. We show that Mlh3p also has a significant role in the repair of four-base loops and demonstrate the existence of a complex that repairs four-base loops and is dependent on Msh2p and Msh3p, but independent of Mlh1p. Finally, we demonstrate that the ratio of the types of repair (conversion vs. restoration) can be altered by changing the level of local double-strand breaks.
MATERIALS AND METHODS
Strains:
All haploid yeast strains were derivatives of AS4 (α arg4-17 trp1 tyr7 ade6 ura3) or AS13 (a leu2 ade6 ura3) (Stapleton and Petes 1991). The genotypes and constructions of haploids and diploids are described in Tables 1 and 2, respectively. All mutant derivatives were constructed by one-step or two-step transplacement or by crosses with isogenic strains. All transformants were verified using PCR analysis. As diploid AS4 × AS4 strains have a sporulation deficiency caused by a mutation within STP22, an STP22-containing plasmid (pDJ173; provided by D. Jenness, University of Massachusetts Medical School) was maintained during AS4 × AS4 crosses. Haploids were cured of pDJ173 prior to use in experiments. pDJ173 was created by inserting the 2-kb HindIII/SalI fragment of pDJ166 (Li et al. 1999) into HindIII/SalI-cut YCp50.
TABLE 1.
Haploid yeast strains
| Strain | Relevant genotypea | Construction details or referenceb |
|---|---|---|
| AS4-derived haploids | ||
| DTK225 | rad1∷ura3 | Kirkpatrick and Petes (1997) |
| HMY104 | msh2∷kanMX4 | Kearney et al. (2001) |
| HMY105 | mlh1∷kanMX4 | Kearney et al. (2001) |
| HMY106 | pms1∷kanMX4 | Kearney et al. (2001) |
| HMY110 | msh3∷kanMX4 | Kearney et al. (2001) |
| HMY118 | exo1∷kanMX4 | Kearney et al. (2001) |
| HMY123 | rad27∷hisG | Kearney et al. (2001) |
| HMY134 | msh6∷kanMX4 | Kearney et al. (2001) |
| HMY158 | mlh3∷kanMX4 | Kearney et al. (2001) |
| HMY161 | mlh2∷kanMX4 | Kearney et al. (2001) |
| HMY195 | mlh1∷hygB his4∷U1.1a | Kearney et al. (2001) |
| HMY223 | msh4∷kanMX4 | Kearney et al. (2001) |
| JSY119 | his4∷ura3 mlh1∷hygB | 5FOAR isolate of HMY195 (Kearney et al. 2001) |
| JSY125 | a + pDJ173 (plasmid-borne STP22 URA3) | Spore colony from MC156 |
| JSY129 | amlh1∷hygB + pDJ173 | Spore colony from JSY119 × JSY125 cross |
| JSY149 | mlh1∷hygB mlh3∷kanMX4 | Spore colony from HMY158 × JSY129 cross |
| JSY181 | mlh1∷hygB mlh2∷kanMX4 | Spore colony from HMY161 × JSY129 cross |
| JSY218 | amsh2∷kanMX4 +pDJ173 | Spore colony from HMY104 × JSY125 cross |
| JSY269 | arg4-tel pms1∷hygB | OST in MW81 with pms1∷hygB, PCR primers f pms1-Δ and r pms1-Δ (Kearney et al. 2001), pA632 template (Goldstein and McCusker 1999) |
| JSY274 | mlh1∷hygB pms1∷kanMX4 | Spore colony from HMY106 × JSY129 cross |
| JSY279 | arg4-tel msh2∷kanMX4 | OST in MW81 with msh2∷kanMX4, PCR primers f msh2-Δ and r msh2-Δ (Kearney et al. 2001), template pFA6-kanM×4 (Wach et al. 1994) |
| JSY293 | msh5∷hygB | OST in AS4 with msh5∷hygB, PCR primers OL65-fMSH5+kan (5′-ACAACTCATTCAAAATAACTTACTCATTCATATACTGCCACCAAATGGAATcg tacgctgcaggtcgac) and OL66-rMSH5+kan (5′-TTATTAACTTAAATATGT TACAAGTAAGCGTTTTTTTATTCTTTGATATAatcgatgaattcgagctcg), pA632 template |
| JSY307 | pol3-01 | TST in AS4 with BamHI-cut YIpAM26 (Morrison et al. 1993) |
| JSY316 | bas1∷hygB | OST in AS4 with bas1∷hygB, PCR primers bas1-del-F (5′-CCGGTTTGCTGATTTGAATCGTTCTCTGCAACAATTGTTGATCTCTAGTGGGG Tcgtacgctgcaggtcgac) and bas1-del-R (5′-GCGAGTCATGAAACTA CACGTTTTTTTCAGAGAATATTATCCATCTTGTGCTatcgatgaattc gagctcg), pA632 template |
| JSY318 | bas1∷hygB msh2∷Tn10LUK7-7 | OST in RKY1721 using primers and template described for JSY316 |
| JSY336 | his4-51 msh2∷kanMX4 | Spore colony from MW30 × JSY218 cross |
| JSY347 | msh2∷kanMX4 msh4∷kanMX4 | Spore colony from JSY218 × HMY223 cross |
| MW30 | his4-51 | White et al. (1991) |
| MW81 | arg4-tel | White et al. (1993) |
| RKY1721 | msh2∷Tn10LUK7-7 | Alani et al. (1994) |
| AS13-derived haploids | ||
| HMY91 | mlh1∷kanMX4 his4-AAG | Welz-Voegele et al. (2002) |
| HMY92 | pms1∷kanMX4 his4-AAG | Welz-Voegele et al. (2002) |
| HMY131 | α | Kearney et al. (2001) |
| JSY127 | α his4-AAG | Spore colony from PD73 x HMY131 (Kearney et al. 2001) cross |
| JSY133 | exo1∷kanMX his4-AAG | Spore colony from HMY119 (Kearney et al. 2001) × JSY127 cross |
| JSY135 | α exo1∷kanMX his4∷U1.1a | Spore colony from HMY119 (Kearney et al. 2001) × JSY127 cross |
| JSY137 | mlh3∷kanMX4 his4-AAG | Spore colony from HMY159 (Kearney et al. 2001) × JSY127 cross |
| JSY139 | mlh2∷kanMX4 his4-AAG | Spore colony from HMY162 (Kearney et al. 2001) × JSY127 cross |
| JSY143 | msh3∷kanMX4 his4-AAG | Spore colony from HMY111 (Kearney et al. 2001) × JSY127 cross |
| JSY144 | msh6∷kanMX4 his4-AAG | Spore colony from HMY135 (Kearney et al. 2001) × JSY127 cross |
| JSY151 | msh4∷kanMX4 his4-AAG | Spore colony from HMY242 (Kearney et al. 2001) × JSY127 cross |
| JSY177 | α HIS4 msh3∷hygB | Spore colony from HMY131 (Kearney et al. 2001) × HMY220 (Kearney et al. 2001) cross |
| JSY183 | α his4-Sal | Spore colony from HMY131 (Kearney et al. 2001) × MW1 cross |
| JSY184 | msh3∷hygB his4-Sal | Spore colony from MW1 × JSY177 cross |
| JSY186 | exo1∷kanMX4 his4-Sal | Spore colony from MW1 × JSY135 cross |
| JSY193 | msh6∷kanMX4 his4-Sal | Spore colony from HMY135 (Kearney et al. 2001) × JSY183 cross |
| JSY195 | mlh1∷hygB his4-Sal | Spore colony from HMY196 (Kearney et al. 2001) × JSY183 cross |
| JSY196 | α mlh1∷hygB his4-Sal | Spore colony from HMY196 (Kearney et al. 2001) × JSY183 cross |
| JSY197 | mlh2∷kanMX4 his4-Sal | Spore colony from HMY162 (Kearney et al. 2001) × JSY183 cross |
| JSY206 | msh2∷kanMX4 his4-Sal | Spore colony from HMY101 (Kearney et al. 2001) × JSY183 cross |
| JSY211 | mlh3∷kanMX4 his4-Sal | Spore colony from HMY159 (Kearney et al. 2001) × JSY183 cross |
| JSY213 | α mlh3∷kanMX4 his4-U1.1a | Spore colony from HMY159 (Kearney et al. 2001) × JSY183 cross |
| JSY214 | α msh2∷kanMX4 his4-AAG | Spore colony from HMY101 (Kearney et al. 2001) × JSY127 cross |
| JSY231 | pms1∷kanMX4 his4-Sal | Spore colony from HMY103 (Kearney et al. 2001) × JSY183 cross |
| JSY234 | rad27∷hisG his4-Sal | Spore colony from HMY145 (Kearney et al. 2001) × JSY183 cross |
| JSY249 | msh4∷kanMX4 his4-Sal | Spore colony from HMY242 (Kearney et al. 2001) × JSY183 cross |
| JSY252 | rad1:ura3 his4-Sal | Spore colony from HMY35 (Kearney et al. 2001) × JSY183 cross |
| JSY259 | mlh1∷hygB mlh2∷kanMX4 his4-Sal | Spore colony from JSY197 × JSY196 cross |
| JSY261 | mlh1∷hygB mlh3∷kanMX4 his4-Sal | Spore colony from JSY196 × JSY211 cross |
| JSY267 | rad27∷hisG his4-Sal | Spore colony from JSY234 × JSY213 cross |
| JSY270 | arg4-tel pms1∷hygB | OST in MW79 with pms1∷hygB, PCR primers f pms1-Δ and r pms1-Δ (Kearney et al. 2001), pA632 template |
| JSY273 | mlh1∷hygB pms1∷kanMX4 his4-Sal | Spore colony from JSY196 × JSY231 cross |
| JSY278 | arg4-tel msh2∷kanMX4 | OST in MW79 with msh2∷kanMX4, PCR primers f msh2-Δ and r msh2-Δ (Kearney et al. 2001), template pFA6-kanMX4 |
| JSY294 | msh5∷hygB his4-AAG | OST in PD73 with msh5∷hygB, PCR primers OL65-fMSH5+kan (5′-ACAACTCATTCAAAATAACTTACTCATTCATATACTGCCACCAAATGGAATcgtacgctgc aggtcgac) and OL66-rMSH5+kan (5′-TTATTAACTTAAATATGTTACAAGTA AGCGTTTTTTTATTCTTTGATATAatcgatgaattcgagctcg), pA632 template |
| JSY295 | msh5∷hygB his4-Sal | OST in MW1 with msh5∷hygB, PCR primers OL65-fMSH5+kan (5′-ACAACTCATTCAAAATAACTTACTCATTCATATACTGCCACCA AATGGAATcgtacgctgcaggtcgac) and OL666-rMSH5+kan (5′-TTATTAACTTAAATATGTTACAAGTAAGCGTTTTTTTATTCTTT GATATAatcgatgaattcgagctcg), pA632 template |
| JSY308 | pol3-01 his4-Sal | TST in MW1 with BamHI-cut YIpAM26 (Morrison et al. 1993) |
| JSY312 | pol3-01 his4-AAG | TST in PD73 with BamHI-cut YIpAM26 (Morrison et al. 1993) |
| JSY317 | bas1∷hygB his4-AAG | OST in PD73 with bas1∷hygB, PCR primers bas1-del-F (5′-CCGGTTTGCTGATTTGAATCGTTCTCTGCAACAATTGTTGATCTCTAGTGGGGTcgtacgctgcag gtcgac) and bas1-del-R (5′-GCGAGTCATGAAACTACACGTTTTTTTCAGA GAATATTATCCATCTTGTGCTatcgatgaattcgagctcg), pA632 template |
| JSY319 | bas1∷hygB msh2∷Tn10LUK7-7 his4-AAG | OST in RKY1452 with bas1∷hygB, PCR primers bas1-del-F (5′-CCGGTTTGCTGATTTGAATCGTTCTCTGCAACAATTGTTGATCTCTAGTGGGGTcgtacgctgc aggtcgac) and bas1-del-R (5′-GCGAGTCATGAAACTACACGTTTTTTTCAGA GAATATTATCCATCTTGTGCTatcgatgaattcgagctcg), pA632 template |
| JSY337 | his4-51 his4-AAG | TST of PD73 with BsrGI-cut pMW35 (White et al. 1991) |
| JSY342 | his4-51 msh2∷kanMX4 | Spore colony from JSY214 × JSY337 cross |
| JSY348 | msh2∷kanMX4 msh4∷kanMX4 | Spore colony from JSY214 × JSY151 cross |
| MW1 | his4-Sal | Nag et al. (1989) |
| MW79 | arg4-tel | White et al. (1993) |
| PD73 | his4-AAG | Detloff et al. (1991) |
| RKY1452 | msh2∷Tn10LUK7-7 his4-AAG | Alani et al. (1994) |
| Mating-type testers | ||
| A364a | aade1 ade2 his5 lys2 ura1 | Petes and Botstein (1977) |
| 2262 | α ade1 his5 leu2 lys1 ura1 | Petes and Botstein (1977) |
All strains, except those for mating-type testers, were derived from the haploid strains AS4 (α arg4-17 trp1 tyr7-1 ade6 ura3) and AS13 (a leu2 ade6 ura3 rme1) (Stapleton and Petes 1991) by transformation or by crosses with isogenic strains. Only those markers that are different from the haploid progenitor strains are shown.
Some strains were made by one-step transplacements (OST) or two-step transplacements (TST). Strains constructed by one-step transplacements were made using PCR fragments containing a selectable gene flanked by sequences of the gene to be deleted (Wach et al. 1994). For each such construction, either the primers or a reference for the primers are indicated as well as the template plasmid. Within the primer sequences, uppercase letters indicate genomic sequences identical to the disrupted gene and lowercase letters indicate sequences identical to the selectable gene.
TABLE 2.
Diploid yeast strains
| Strain | Relevant homozygous mutations | Crossa |
|---|---|---|
| Strains heterozygous for his4-AAG | ||
| DTK224 | rad1∷ura3 | TP1011 × TP1010 (Kirkpatrick and Petes 1997) |
| HMY95 | mlh1∷kanMX4 | HMY105 × HMY91 (Welz-Voegele et al. 2002) |
| HMY96 | pms1∷kanMX4 | DNY95 × HMY92 (Welz-Voegele et al. 2002) |
| JSY136 | exo1∷kanMX4 | HMY118 × JSY133 |
| JSY163 | msh3∷kanMX4 | HMY110 × JSY143 |
| JSY164 | msh6∷kanMX4 | HMY134 × JSY144 |
| JSY165 | mlh2∷kanMX4 | HMY161 × JSY139 |
| JSY178 | mlh3∷kanMX4 | HMY158 × JSY137 |
| JSY291 | msh4∷kanMX4 | HMY223 × JSY151 |
| JSY297 | msh5∷hygB | JSY293 × JSY294 |
| JSY314 | pol3-01 | JSY307 × JSY312 |
| JSY320 | bas1∷hygB | JSY316 × JSY317 |
| JSY321 | bas1∷hygB msh2∷Tn10LUK7-7 | JSY318 × JSY319 |
| JSY338 | his4-51 | MW30 × JSY337 |
| JSY343 | his4-51 msh2∷kanMX4 | JSY336 × JSY342 |
| JSY349 | msh2∷kanMX4 msh4∷kanMX4 | JSY347 × JSY348 |
| No. 5 | msh2∷Tn10LUK7-7 | RJK1721 × RJK 1452 (Alani et al. 1994) |
| PD83 | Wild type | AS4 × PD73 (Detloff et al. 1991) |
| Strains heterozygous for his4-Sal | ||
| JSY188 | msh3∷hygB/kanMX4 | HMY110 × JSY184 |
| JSY210 | mlh2∷kanMX4 | HMY161 × JSY197 |
| JSY216 | mlh3∷kanMX4 | HMY158 × JSY211 |
| JSY217 | msh6∷kanMX4 | HMY134 × JSY193 |
| JSY230 | mlh1∷hygB/kanMX4 | HMY105 × JSY195 |
| JSY233 | pms1∷kanMX4 | HMY106 × JSY231 |
| JSY239 | rad27∷hisG | HMY123 × JSY267 |
| JSY240 | msh2∷kanMX4 | HMY104 × JSY206 |
| JSY255 | rad1:ura3 | DTK225 × JSY252 |
| JSY277 | mlh1∷hygB pms1∷kanMX4 | JSY274 × JSY273 |
| JSY283 | mlh1∷hygB mlh3∷kanMX4 | JSY149 × JSY261 |
| JSY284 | exo1∷kanMX4 | HMY118 × JSY186 |
| JSY285 | msh4∷kanMX4 | HMY223 × JSY249 |
| JSY292 | mlh1∷hygB mlh2∷kanMX4 | JSY181 × JSY259 |
| JSY296 | msh5∷hygB | JSY293 × JSY295 |
| JSY313 | pol3-01 | JSY307 × JSY308 |
| MW103 | Wild type | AS4 × MW1 (Nag et al. 1989) |
| Other diploids | ||
| JSY276 | arg4-tel pms1∷hygB | JSY269 × JSY270 |
| JSY282 | arg4-tel msh2∷kanMX4 | JSY279 × JSY278 |
| MW160 | arg4-tel | MW81 × MW79 (White et al. 1993) |
| MC153 | Wild type | AS4 × AS4 [created by inducing HO expression on the plasmid pGAL-HO (Herskowitz and Jensen 1991) in AS4 and then selecting for a diploid that had lost the plasmid] |
| MC156 | Wild type +pDJ173, plasmid-borne STP22 URA3 | MW153, carrying pDJ173 (Li et al. 1999) |
Genotypes of the haploids used in the constructions are given in Table 1. In crosses, AS4- and AS13-derived strains are shown at the left and the right of the ×, respectively.
Genetic techniques:
Standard media and genetic methods were used (Sherman et al. 1982) except where indicated. Some of the mutant strains examined have mutator phenotypes, resulting in increased levels of spore inviability. To maximize spore viability, we generated diploids by mating haploid strains overnight and sporulating the diploids on the following day without purification. As in previous studies, diploids were sporulated at 18° to maximize meiotic recombination at HIS4 (Fan et al. 1995). Following tetrad dissection on plates containing rich growth media (YPD), we replica plated the resulting spore colonies to various omission media to score segregating markers. Spore colonies from strains carrying the his4-Sal mutation were examined microscopically for small sectors on both media lacking histidine and media lacking arginine. Spore colonies from strains carrying the his4-AAG allele, with a few exceptions, were examined microscopically only on medium lacking histidine.
To detect chromosome III nondisjunction events, spore colonies were replica plated to tester strains of the a- and α-mating types. After overnight incubation, the mated cells were replica plated to omission medium lacking adenine. Tetrads were considered to have meiosis I nondisjunction events involving chromosome III when only two spores were viable and both lacked the ability to mate. This procedure will underestimate the frequency of nondisjunction, if the nondisjunction events are unrelated to a lack of meiotic crossovers.
Statistical analysis:
Comparisons were made using the Fisher exact test with two-tail P-values or by chi-square analysis (for comparisons involving more than two experimental parameters or comparisons with numbers too large for the Fisher exact test) using the statistical analysis program on the VassarStats website (http://faculty.vassar.edu/lowry/vassarstats.html). Whether one- or two-tailed tests were done is indicated in Tables 3–6. Because of the large numbers of comparisons performed, we used the sequential P-value procedure (Benjamini and Hochberg 1995) to limit the false discovery rate. For each set of comparisons, we first determined the uncorrected P-values and then used the Benjamini–Hochberg procedure with an initial value of α = 0.05 to determine which P-values were statistically significant.
TABLE 3.
Meiotic segregation patterns for the his4-AAG marker
| % of total tetrads
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Relevant genotype | No. of tetrads | Aberrant segregationa (%) | PMSb (%) | PMS/aberrant eventsc (%) | 6:2 | 2:6 | 5:3 | 3:5 | Other aberrant eventsd |
| PD83e | Wild type | 482 | 58 | 10 | 18 | 15 | 22 | 5 | 4 | 11 |
| No. 5f | msh2Δ | 111 | 64 | 55 | 90g | 5 | 3 | 14 | 23 | 19 |
| JSY163 | msh3Δ | 253 | 59 | 10 | 18 | 18 | 18 | 4 | 3 | 17 |
| JSY291 | msh4Δ | 95 | 41h | 7 | 21 | 19 | 12 | 2 | 3 | 5 |
| JSY297 | msh5Δ | 119 | 47h | 9 | 18 | 14 | 20 | 2 | 8 | 3 |
| JSY164 | msh6Δ | 208 | 57 | 46 | 85g | 5 | 4 | 16 | 16 | 16 |
| JSY349 | msh2Δ msh4Δ | 198 | 45h | 40 | 91g | 4 | 1 | 16 | 13 | 12 |
| HMY95e | mlh1Δ | 465 | 57 | 49 | 90g | 4 | 3 | 18 | 15 | 17 |
| JSY165 | mlh2Δ | 194 | 42h | 8 | 23 | 12 | 18 | 4 | 3 | 6 |
| JSY178 | mlh3Δ | 155 | 65 | 11 | 18 | 17 | 22 | 8 | 3 | 16 |
| HMY96e | pms1Δ | 641 | 59 | 43 | 78g | 5 | 6 | 15 | 16 | 16 |
| JSY136 | exo1Δ | 209 | 30h | 9 | 30 | 9 | 10 | 3 | 5 | 3 |
| JSY314 | pol3-01 | 245 | 38h | 2 | 9g | 17 | 15 | 1 | 1 | 4 |
| DTK224f | rad1Δ | 220 | 64h | 13 | 21 | 19 | 22 | 6 | 5 | 12 |
| JSY320 | bas1Δ | 285 | 11h | 3 | 26 | 4 | 5 | 2 | 1 | 0 |
| JSY321 | bas1Δ msh2Δ | 220 | 21hi | 16 | 77g | 3 | 2 | 7 | 7 | 2 |
| JSY338 | his4-51 | 321 | 13h | 3 | 26 | 3 | 7 | 1 | 1 | 1 |
| JSY343 | his4-51 msh2Δ | 316 | 15h | 12 | 81g | 1 | 2 | 5 | 5 | 2 |
All diploids were made by batch mating (described in materials and methods) except DTK224, which is a purified diploid. All strains were sporulated at 18°.
Percentage of total tetrads with aberrant (non-4:4) segregation.
Percentage of total tetrads with one or more PMS events, excluding tetrads with one gene conversion event and one PMS event (7:1, 1:7).
Percentage of aberrant events that are PMS events, calculated as described in the text.
This class includes tetrads with two or more PMS (ab4:4, ab6:2, ab2:6, dev5:3, dev3:5, dev4:4) events, one PMS event and one conversion event (7:1, 1:7), and two conversion events (8:0, 0:8).
Data from Welz-Voegle et al. (2002).
Data from Kirkpatrick and Petes (1997).
Significant difference in PMS events relative to gene conversion events compared to the wild-type strain (PD83). The P-values were determined using two-tailed Fisher exact tests. The P-values for each of the 17 comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.024 were considered significant.
Frequency of aberrant segregation significantly different from that of wild type (PD83). The P-values were determined using two-tailed Fisher exact tests. The P-values for each of the 17 comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.032 were considered significant.
Frequency of aberrant segregation in JSY321 (bas1Δ msh2Δ) significantly increased compared to the bas1Δ strain (JSY320). A one-tailed Fisher exact test was used to determine the P-value of 0.0041.
TABLE 4.
Meiotic segregation patterns for the arg4-17 marker
| % of total tetrads
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain(s) | Relevant genotype | No. of tetrads | Aberrant segregation (%) | PMS (%) | PMS/aberrant events (%) | 6:2 | 2:6 | 5:3 | 3:5 | Other aberrant events |
| MW103/JSY338a | Wild type | 742 | 8 | 0 | 2 | 4 | 3 | 0 | 0 | 1 |
| JSY240/JSY343a | msh2Δ | 502 | 16b | 11 | 67c | 4 | 1 | 5 | 4 | 2 |
| JSY188 | msh3Δ | 167 | 8 | 0 | 0 | 5 | 4 | 0 | 0 | 0 |
| JSY285 | msh4Δ | 224 | 5 | 0 | 0 | 4 | 2 | 0 | 0 | 0 |
| JSY296 | msh5Δ | 155 | 6 | 0 | 0 | 3 | 3 | 0 | 0 | 0 |
| JSY217 | msh6Δ | 182 | 17b | 5 | 61c | 5 | 1 | 7 | 2 | 2 |
| JSY349 | msh2Δ msh4Δ | 198 | 17b | 13 | 83c | 4 | 1 | 4 | 8 | 2 |
| JSY230 | mlh1Δ | 193 | 13b | 7 | 50c | 4 | 2 | 3 | 4 | 1 |
| JSY210 | mlh2Δ | 190 | 8 | 0 | 0 | 5 | 3 | 0 | 0 | 0 |
| JSY216 | mlh3Δ | 219 | 10 | 0 | 0 | 7 | 2 | 0 | 0 | 0 |
| JSY233 | pms1Δ | 159 | 9 | 4 | 43c | 3 | 3 | 3 | 1 | 0 |
| JSY277 | mlh1Δ pms1Δ | 383 | 14b | 8 | 54c | 4 | 3 | 3 | 4 | 1 |
| JSY292 | mlh1Δ mlh2Δ | 181 | 12 | 4 | 29c | 2 | 6 | 2 | 1 | 1 |
| JSY283 | mlh1Δ mlh3Δ | 388 | 15b | 9 | 63c | 4 | 2 | 3 | 6 | 0 |
| JSY284 | exo1Δ | 196 | 4 | 0 | 0 | 1 | 3 | 0 | 0 | 0 |
| JSY313 | pol3-01 | 173 | 9 | 0 | 0 | 5 | 2 | 0 | 0 | 1 |
| JSY255 | rad1Δ | 202 | 11 | 1 | 4 | 5 | 3 | 0 | 0 | 1 |
| JSY239 | rad27Δ | 242 | 8 | 0 | 0 | 3 | 5 | 0 | 0 | 0 |
| MW160 | arg4-teld | 291 | 45b | 1 | 1 | 24 | 18 | 0 | 1 | 3 |
| JSY282 | arg4-teldmsh2Δ | 276 | 51b | 40 | 81c | 8 | 2 | 16 | 17 | 8 |
| JSY276 | arg4-teldpms1Δ | 252 | 46b | 28 | 64c | 12 | 5 | 13 | 12 | 5 |
| JSY320 | bas1Δ | 285 | 6 | 0 | 0 | 3 | 3 | 0 | 0 | 0 |
| JSY321 | bas1Δ msh2Δ | 220 | 23b | 14 | 57c | 6 | 2 | 8 | 5 | 1 |
All diploids were made by batch mating and were sporulated at 18°. The column headings have the same definitions as those of Table 3.
Data from these two strains were pooled. The two strains are isogenic except for the his4-51 mutation, which has no significant effect on segregation of the arg4-17 marker.
Frequency of aberrant segregation significantly different from the wild-type strains MW103 and JSY338. The P-values were determined using two-tailed Fisher exact tests. The P-values for each of the 22 comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.023 were considered significant.
Significant increase in PMS events relative to gene conversion events compared to wild-type MW103 and JSY338. The P-values were determined using one-tailed Fisher exact tests. The P-values for each of the 22 comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.025 were considered significant.
arg4-tel is a homozygous insertion of 60 bp of telomeric sequence upstream of ARG4.
TABLE 5.
Meiotic segregation patterns for the his4-Sal marker
| % of total tetrads
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Relevant genotype | No. of tetrads | Aberrant segregation (%) | PMS (%) | PMS/aberrant events (%) | 6:2 | 2:6 | 5:3 | 3:5 | Other aberrant events |
| MW103 | Wild type | 421 | 25 | 0 | 0 | 10 | 13 | 0 | 0 | 3 |
| JSY240 | msh2Δ | 186 | 37 | 30 | 84b | 4 | 3 | 12 | 11 | 6 |
| JSY188 | msh3Δ | 167 | 47a | 34 | 76b | 9 | 2 | 16 | 11 | 9 |
| JSY285 | msh4Δ | 224 | 17a | 0 | 0 | 8 | 9 | 0 | 0 | 0 |
| JSY296 | msh5Δ | 155 | 22 | 0 | 0 | 7 | 15 | 0 | 0 | 1 |
| JSY217 | msh6Δ | 182 | 26 | 0 | 0 | 12 | 12 | 0 | 0 | 2 |
| JSY230 | mlh1Δ | 193 | 37 | 19 | 54bc | 14 | 2 | 10 | 6 | 5 |
| JSY210 | mlh2Δ | 190 | 23 | 0 | 0 | 7 | 13 | 0 | 0 | 3 |
| JSY216 | mlh3Δ | 219 | 27 | 7 | 28bcd | 11 | 7 | 3 | 4 | 2 |
| JSY233 | pms1Δ | 159 | 31 | 18 | 59bc | 7 | 4 | 8 | 8 | 3 |
| JSY277 | mlh1Δ pms1Δ | 383 | 25 | 16 | 67b | 6 | 3 | 9 | 6 | 2 |
| JSY292 | mlh1Δ mlh2Δ | 181 | 28 | 15 | 55b | 9 | 3 | 7 | 7 | 1 |
| JSY283 | mlh1Δ mlh3Δ | 388 | 30 | 21 | 70be | 6 | 2 | 11 | 8 | 3 |
| JSY284 | exo1Δ | 196 | 19a | 1 | 3 | 10 | 8 | 0 | 1 | 1 |
| JSY313 | pol3-01 | 173 | 17a | 0 | 0 | 7 | 9 | 0 | 0 | 1 |
| JSY255 | rad1Δ | 202 | 23 | 0 | 0 | 8 | 15 | 0 | 0 | 1 |
| JSY239 | rad27Δ | 242 | 21a | 0 | 0 | 10 | 11 | 0 | 0 | 0 |
All diploids were made by batch mating and were sporulated at 18°. The column headings have the same definitions as those of Table 3.
Frequency of aberrant segregation significantly different from wild type (MW103). The P-values were determined using two-tailed Fisher exact tests. The P-values for each of the 16 comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.0125 were considered significant.
Significant increase in PMS events relative to gene conversion events compared to the wild-type strain (MW103). The P-values were determined using one-tailed Fisher exact tests. The P-values for each of the 16 comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.025 were considered significant.
Significant decrease in PMS events relative to gene conversion events compared to the msh2Δ strain (JSY240). The P-values were determined using one-tailed Fisher exact tests. The P-values for three comparisons (JSY240 vs. JSY230, JSY216, and JSY233) were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.05 were considered significant.
Significant decrease in PMS events relative to gene conversion events compared to the mlh1Δ strain (JSY230) and to the pms1Δ (JSY233) strain. The P-values were determined using one-tailed Fisher exact tests. The P-values for two comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.05 were considered significant.
Significant increase in PMS events relative to gene conversion events compared to the mlh1Δ strain (JSY230). These events were compared by one-tailed Fisher exact tests using data from the strains JSY277 (mlh1Δ pms1Δ), JSY292 (mlh1Δ mlh2Δ), and JSY283 (mlh1Δ mlh3Δ). The P-values for the resulting three comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.017 were considered significant.
TABLE 6.
Genetic distances in three genetic intervals on chromosome III
|
HIS4–LEU2
|
LEU2–CEN3
|
CEN3–MAT
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain(s)a | Relevant genotype | P | N | T | CMb | P | N | T | CMc | P | N | T | CMc |
| PD83/MW103 | Wild type | 286 | 8 | 168 | 24 | 358 | 338 | 165 | 10 | 209 | 185 | 364 | 24 |
| JSY240 | msh2Δ | 73 | 3 | 37 | 24 | 70 | 74 | 33 | 9 | 40 | 45 | 82 | 25 |
| JSY163/JSY188 | msh3Δ | 104 | 4 | 81 | 28 | 157 | 162 | 78 | 10 | 91 | 95 | 168 | 24 |
| JSY291/JSY285 | msh4Δ | 172 | 2 | 49 | 14d | 129 | 113 | 47 | 8 | 107 | 112 | 88 | 14e |
| JSY297/JSY296 | msh5Δ | 141 | 0 | 31 | 9d | 112 | 106 | 40 | 8 | 106 | 99 | 55 | 11e |
| JSY164/JSY216 | msh6Δ | 120 | 3 | 87 | 25 | 132 | 161 | 78 | 11 | 70 | 82 | 154 | 25 |
| JSY349 | msh2Δ msh4Δ | 90 | 1 | 17 | 11d | 81 | 83 | 33 | 8 | 76 | 71 | 40 | 11e |
| HMY95/JSY230 | mlh1Δ | 220 | 3 | 77 | 16d | 261 | 258 | 125 | 10 | 142 | 155 | 121 | 14e |
| JSY165/JSY210 | mlh2Δ | 155 | 5 | 87 | 24 | 138 | 168 | 61 | 8 | 97 | 108 | 162 | 22 |
| JSY178/JSY216 | mlh3Δ | 133 | 4 | 81 | 24 | 168 | 155 | 50 | 7 | 148 | 123 | 140 | 17e |
| HMY96/JSY233 | pms1Δ | 226 | 1 | 128 | 19 | 325 | 336 | 117 | 8 | 136 | 116 | 192 | 22 |
| JSY277 | mlh1Δ pms1Δ | 210 | 0 | 70 | 13d | 159 | 147 | 62 | 8 | 127 | 102 | 108 | 16e |
| JSY292 | mlh1Δ mlh2Δ | 99 | 0 | 25 | 10d | 60 | 75 | 33 | 10 | 53 | 75 | 46 | 13e |
| JSY283 | mlh1Δ mlh3Δ | 180 | 3 | 74 | 18 | 130 | 163 | 70 | 10 | 120 | 142 | 117 | 15e |
| JSY136/JSY284 | exo1Δ | 223 | 2 | 69 | 14d | 170 | 166 | 30 | 4f | 132 | 139 | 84 | 12e |
| JSY314/JSY313 | pol3-01 | 85 | 5 | 43 | 27 | 55 | 63 | 38 | 12 | 35 | 43 | 77 | 25 |
| JSY255 | rad1Δ | 82 | 7 | 56 | 34 | 85 | 91 | 23 | 6 | 74 | 46 | 79 | 20 |
| JSY239 | rad27Δ | 127 | 2 | 54 | 18 | 96 | 93 | 39 | 9 | 36 | 36 | 77 | 26 |
| JSY320 | bas1Δ | 163 | 1 | 63 | 15 | 93 | 95 | 54 | 11 | 67 | 75 | 110 | 22 |
| JSY321 | bas1Δ msh2Δ | 108 | 1 | 45 | 17 | 77 | 78 | 41 | 11 | 55 | 47 | 86 | 23 |
| JSY338 | his4-51 | 195 | 5 | 65 | 18d | 130 | 109 | 53 | 9 | 101 | 69 | 135 | 22 |
| JSY343 | his4-51 msh2Δ | 184 | 11 | 102 | 29 | 131 | 100 | 74 | 12 | 78 | 70 | 151 | 25 |
P, N, and T indicate parental ditype (PD), nonparental ditype (NPD), and tetratype tetrads (T), respectively. We included only those tetrads in which both flanking markers had Mendelian segregation.
For some intervals, data from multiple strains were pooled. These strains were isogenic, except for the specific mutations at the HIS4 locus.
Calculated by the equations of Perkins (1949).
Calculated as the percentage of second-division segregation (tetratype tetrads) divided by two. First-division segregation (FDS) and second-division segregation (SDS) tetrads were determined using the heterozygous centromere-linked trp1 marker.
Significant difference compared to the wild-type strain (PD83) in the numbers of PD:NPD:T tetrads. The P-values were determined using chi-square analysis. The P-values for each of the 21 comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.019 were considered significant.
Significant difference compared to the wild-type strain (PD83) in the numbers of FDS:SDS tetrads for the CEN3–MAT interval. The P-values were determined using two-tailed Fisher exact tests. The P-values for 21 comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.021 were considered significant.
Significant difference compared to the wild-type strain (PD83) in the numbers of FDS:SDS tetrads for the LEU2–CEN3 interval. The P-values were determined using two-tailed Fisher exact tests. The P-values for 21 comparisons were ranked and the Benjamini and Hochberg (1995) procedure was applied using an initial α = 0.05. P-values <0.002 were considered significant.
RESULTS
Experimental system:
To examine the roles of the individual MMR proteins in the repair of different types of DNA substrates, we used isogenic diploid S. cerevisiae strains heterozygous for either the his4-AAG or the his4-Sal alleles. The his4-AAG allele is a T-to-A substitution at the second position of the HIS4 start codon. A heteroduplex formed between a strand with wild-type information and one with the his4-AAG substitution will result in an A/A or T/T mismatch, depending on which strand is transferred in forming the heteroduplex (Detloff et al. 1991). We chose to examine this particular mutant substitution because our diploids are also heterozygous for the arg4-17 allele, which contains a T-to-A mutation at position +127 of the ARG4 coding sequence (White et al. 1985). Thus, by using isogenic strains containing the his4-AAG and the arg4-17 alleles, we are able to determine whether the same mismatch in different contexts is repaired in the same manner. The his4-Sal allele is a four-base restriction site fill-in located at position +467 of the HIS4 coding sequence (Nag et al. 1989). A heteroduplex formed by one strand containing the wild-type HIS4 sequences and one strand containing the his4-Sal sequence results in a four-base loop.
In our genetic background, the HIS4 locus undergoes a very high rate of meiotic recombination as a consequence of a very strong double-strand break (DSB) located ∼250 bp upstream of the HIS4 initiation codon (Fan et al. 1995; Xu and Petes 1996). The high rate of DSB formation results in high rates of heteroduplex formation involving markers within the HIS4 gene. In a strain heterozygous for the his4-AAG mutation, a DSB on the wild-type or mutant chromosomes results in an A/A or a T/T mismatch, respectively. If the heteroduplex reflects a DSB on the wild-type strand, failure to correct the resulting mismatch results in a tetrad with one His+, two His−, and one His+/− sectored spore colonies; such tetrads are termed “3:5,” following the nomenclature used for eight-spored fungi. Repair of the mismatch either can yield a gene conversion (2:6) or can restore Mendelian segregation (4:4). Similarly, an event initiated on the mutant chromosome will generate a 5:3 or 6:2 tetrad or restore Mendelian segregation. Since the HIS4 locus has such a high level of recombination, we also find tetrads with more than one aberrant segregation event. For example, a tetrad with three wild-type spore colonies and one sectored colony is termed a 7:1 segregation event and assumed to reflect a tetrad with one gene conversion event and one postmeiotic segregation (PMS) event (Nag et al. 1989). Tetrads with more than one aberrant segregation event are usually <10% as frequent as those with one aberrant segregation event.
In many studies, the efficiency of repair of mismatches formed during meiotic recombination is estimated by dividing the number of tetrads with one or more PMS events by the total number of aberrant segregation events (tetrads with one or more PMS events, one or more gene conversion events, and tetrads with both gene conversion and PMS events). In this study, we use a different calculation that gives the appropriate weight to tetrads with two or more aberrant segregation events. More specifically, we calculate the percentage of PMS events per aberrant events by summing the numbers of tetrads with a single PMS spore colony (5:3 + 3:5 + 7:1 + 1:7), two times the number of tetrads with two PMS spore colonies (aberrant 4:4 + aberrant 6:2 + aberrant 2:6), three times the number of tetrads with three PMS spore colonies (deviant 5:3 + deviant 3:5), and four times the number of tetrads with four PMS spore colonies (deviant 4:4). The resulting sum is divided by the sum of aberrant events, calculated by summing the number of tetrads with one aberrant event (6:2 + 2:6 + 5:3 + 3:5), two times the number of tetrads with two aberrant events (7:1 + 1:7 + 8:0 + 0:8 + aberrant 4:4 + aberrant 6:2 + aberrant 2:6), three times the number of tetrads with three aberrant events (deviant 5:3 + deviant 3:5), and four times the number of tetrads with four aberrant events (deviant 4:4). The aberrant segregation classes for tetrads with two or more events are defined in Detloff et al. (1991). The tetrads with three or more aberrant events represent <10% of the total aberrant tetrads in all strains examined in this study.
Effects of mutations in MutS and MutL homologs on meiotic MMR of the base–base mismatch:
As expected from previous studies, the heterozygous his4-AAG mutation had a very high rate of aberrant segregation—∼60% of the tetrads of the wild-type strain (PD83; Table 3). For most genetic loci in yeast, the frequency of aberrant segregation is 2–10% (Petes et al. 1991). In the wild-type strain, repair of the A/A, T/T mismatch at the HIS4 hotspot is not complete, since 18% of the aberrant segregation events are PMS. Interestingly, for the same mismatch at the ARG4 locus, repair is complete in the MMR-proficient MW103 strain (Table 4), arguing that the efficiency of MMR is somewhat context dependent. It should be noted that most of the data for the arg4-17 mismatch came from strains heterozygous for the his4-Sal allele.
The effects of the mutations in the MutS and MutL homologs on the repair of the base–base mismatch can be divided into two groups. Deletions of MSH2, MSH6, MLH1, and PMS1 had similar effects, resulting in ∼80–90% PMS/aberrant events for his4-AAG. Deletions of MSH3, MSH4, MSH5, MLH2, and MLH3 did not significantly affect the ratio of PMS/aberrant events. These results are consistent with the conclusion that most of the meiotic repair events for base–base mismatches are initiated by a complex of the Msh2/Msh6/Mlh1/Pms1 proteins. Similar results were observed for the arg4-17 marker, although the percentages of PMS/aberrant events are generally lower (45–83%) relative to those seen for the his4-AAG marker. Thus, some gene conversion events occur by a mechanism that is independent of the standard MMR pathway, and this mechanism is more efficient at the arg4-17 site than at the his4-AAG site.
Strains with mutations in msh4, msh5, and exo1 had significantly reduced levels of aberrant segregation of the his4-AAG marker (Table 3). The exo1 mutants have been previously reported to reduce the frequency of aberrant segregation (Khazanehdari and Borts 2000; Kirkpatrick et al. 2000; Abdullah et al. 2004). Other researchers found that deletion of msh4 or msh5 has little effect on the frequency of aberrant segregation for most markers, although the arg4-Nsp allele was reported to have an elevated frequency of aberrant segregation in msh4 strains (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995). The reduction in aberrant segregation observed in our study could reflect either a reduced frequency of heteroduplexes that include his4-AAG or an increase in the frequency of restoration-type repair relative to conversion-type repair. A reduction in heteroduplex formation by the msh4 mutation would be expected to reduce the rate of aberrant segregation in a MMR-deficient strain, whereas increased restoration-type repair would have relatively little effect in such a strain. We found a significant (P = 0.002 by one-tailed Fisher exact test) reduction in aberrant segregation in the msh2 msh4 mutant compared to the msh2 mutant. The level of aberrant segregation was not significantly different in the msh2 msh4 mutant compared to the msh4 single mutant. These results argue that the level of heteroduplexes that include the his4-AAG mismatch is reduced in the msh4 strain. If Msh4p/Msh5p-promoted crossovers contribute to heteroduplex formation at the his4-AAG mismatch, the observed reduction in aberrant segregation is predicted.
Effects of mutations in MutS and MutL homologs on meiotic MMR of a four-base loop:
The meiotic repair of the four-base loop was examined in diploids with the his4-Sal allele (Table 5). First, as expected from previous studies (reviewed by Surtees et al. 2004), the msh6 mutation had no effect on loop repair, and the msh3 mutation had approximately the same effect as msh2; this result is consistent with the previous conclusion that most of the repair of the four-base loop involves a heterodimer of Msh2p and Msh3p. Second, the effects of the mlh1 and pms1 on loop repair were significantly weaker than the effect of msh2. Third, the mlh3 mutation significantly affected the repair of the four-base loop, although the effect was significantly less than that observed for mlh1 or pms1.
To examine further the roles of the various MutL homologs in the repair of the four-base loop, we also examined the efficiency of repair in various double-mutant strains. Of the mlh1 pms1, mlh1 mlh2, and mlh1 mlh3 double mutants, only the mlh1 mlh3 double mutant had a significantly greater repair defect than mlh1 (Table 5). The simplest interpretation of these data (to be discussed further below) is that most of the repair of the four-base loops is dependent on a heterodimer of Mlh1p and Pms1p, but some fraction of the repair events require Mlh3p and do not require Mlh1p.
Effects of mutations in genes encoding nucleases on meiotic MMR:
As discussed in the Introduction, Exo1p has a role in the excision step of MMR, but other nucleases that are partially redundant with Exo1p are also involved. Candidates for these nucleases include Rad1p/Rad10p, Rad27p, and the proofreading activity of DNA polymerase δ. We previously showed that Rad1p and Rad10p were involved in the meiotic repair of 26-base and 1-kb loops, but not in the repair of a base–base mismatch or a four-base loop (Kirkpatrick and Petes 1997; Kearney et al. 2001). Since these experiments were done under slightly different conditions from those employed in this study, we repeated this analysis for the his4-Sal and arg4-17 markers.
As shown in Tables 3–5, there was no significant effect of exo1, rad1, rad27, or the pol3-01 (encoding a proofreading exonuclease-deficient DNA polymerase δ) on the efficiency of DNA mismatch repair of the base–base mismatch or the four-base loop. These results confirm previous studies of exo1 (Khazanehdari and Borts 2000; Kirkpatrick et al. 2000; Tsubouchi and Ogawa 2000) and rad1 (Kirkpatrick and Petes 1997). Although we did not examine the effect of the rad27 mutation in diploids heterozygous for his4-AAG, the mutation had no effect on the repair of the same mismatch (A/A, T/T) at the ARG4 locus (Table 4). The lack of effect of these nucleases on the efficiency of meiotic MMR argues that they have no function in meiotic MMR or that their functions are redundant. Unfortunately, the lethality of some double-mutant combinations of these nuclease-encoding genes (for example, rad27 pol3-01; Kokoska et al. 1998) precludes a complete analysis of this issue.
In pol3-01 strains, although the efficiency of MMR is not decreased, the rates of aberrant segregation for both types of his4 alleles were significantly reduced (Tables 3 and 5). Maloisel et al. (2004) recently reported that another pol3 mutation (pol3-ct) reduces aberrant segregation and crossovers, without having an effect on mitotic DNA replication. The authors suggest that DNA synthesis by DNA polymerase δ is required for both the elongation of the strand-exchange intermediate and the decision to resolve recombination intermediates as crossovers. In addition to these possibilities, the pol3-01 mutation could increase the relative frequency of restoration-type repair compared to conversion-type repair or alter the level of DSBs. Although we observed no effect of pol3-01 on crossovers (arguing against the last explanation), the level of crossovers is not necessarily directly related to the frequency of DSBs (Henderson and Keeney 2004).
Reduced meiotic crossovers in msh4, msh5, mlh1, mlh3, and exo1 mutants:
We measured crossovers for all strains in the study in three genetic intervals on chromosome III (Table 6). Most of the mutants had no significant effect on crossing over in these intervals. On the basis of genetic and physical studies, it has been suggested that meiotic crossovers are promoted by Msh4p, Msh5p, Mlh1p, and Mlh3p (Hunter and Borts 1997; Wang et al. 1999; Kirkpatrick et al. 2000; Argueso et al. 2004; Börner et al. 2004). In support of this conclusion, we found a significant reduction in crossovers in the CEN3–MAT interval for strains with mutations in the genes encoding these four proteins (Table 6). In addition, we found that crossovers were reduced in the exo1 mutant strains, as expected from previous studies (reviewed by Hoffmann and Borts 2004).
As increased levels of nondisjunction are often associated with reductions in crossovers, we monitored meiosis I nondisjunction genetically (as described in materials and methods). Data from both strains of each genotype were pooled together to determine the ratio of tetrads with nondisjunction of chromosome III to total tetrads: wild type, 0/1665; msh4, 3/464; msh2 msh4, 2/569; msh5, 10/403; mlh1, 2/1585; mlh3, 1/683; mlh1 mlh3, 3/906; mlh1 pms1, 8/575; and exo1, 2/621. No nondisjunction events were observed in any of the other strains used in this study. Of the eight strains with nondisjunction events, only the msh4, msh5, and mlh1 pms1 strains had a significant increase in nondisjunction compared to the wild type. The statistical analysis involved one-tailed chi-square tests with P-values corrected by the Benjamini–Hochberg method; P-values <0.019 were considered significant. The relatively low numbers of nondisjunction events involving chromosome III found in our study, compared to other studies (Hunter and Borts 1997; Wang et al. 1999), may be due to the presence of the very strong HIS4 hotspot on chromosome III in our genetic background.
Conversion-type and restoration-type MMR for the A/A, T/T mismatch:
As discussed above, DNA mismatches can be repaired to generate a gene conversion event or to restore Mendelian segregation. We have previously argued that an efficiently repaired base–base mismatch located near the 5′-end of HIS4 (and, therefore, near the DSB site that initiates recombination) is usually repaired to yield a gene conversion event (Detloff et al. 1992). This conclusion was based on the observation that the aberrant segregation frequency of an efficiently repaired mismatch located near the 5′-end of the gene was approximately the same as that of an inefficiently repaired mismatch. The argument that led to this conclusion is best explained by an example. If an efficiently repaired mismatch occurs in 50% of the tetrads and is repaired equally frequently by conversion-type and restoration-type repair, then the observed frequency of aberrant segregation for this mismatch will be 25%. If an inefficiently repaired mismatch at the same location occurs in 50% of the tetrads, one should observe 50% aberrant segregation. Thus, if a difference in the frequency of aberrant segregation is observed for efficiently and inefficiently repaired mismatches at the same position in the gene, one can infer restoration-type repair; a lack of a difference suggests a lack of restoration-type repair.
From the data shown in Table 3, we can make a similar argument. If we denote the frequency of heteroduplex formation generating a mismatch by H, the frequency of failure to repair the mismatch by NR, the frequency of conversion-type repair as CR, and the frequency of restoration-type repair as RR, then H = NR + CR + RR. At the HIS4 locus in the wild-type strain PD83, since 10% of the tetrads were PMS (NR tetrads) and 48% were conversions (CR tetrads), H(wild type) = 10% + 48% + RR(wild type). In the msh2 strain, the comparable equation is: H(msh2) = 58% + 6% + RR(msh2). Since heteroduplex frequency is the same in the wild-type and msh2 strains (Vedel and Nicolas 1999), H(wild type) = H(msh2). Thus, RR(wild type) = [(58% + 6% + RR(msh2)) − (10% + 48%)]. If we assume that the frequency of restoration-type repair in the absence of msh2 is negligible, then we estimate the percentage of restoration-type repair as 6%, a value much lower than the measured rate of conversion-type repair in the same strain (48%). This result is shown in Figure 1A. In contrast, in isogenic strains, the same calculations indicate that the arg4-17 marker has a level of restoration-type repair equal to the level of conversion-type repair in the wild-type strain. The frequency of conversion-type repair is 7% (Table 4) and the calculated frequency of restoration-type repair is 9% (Figure 1B).
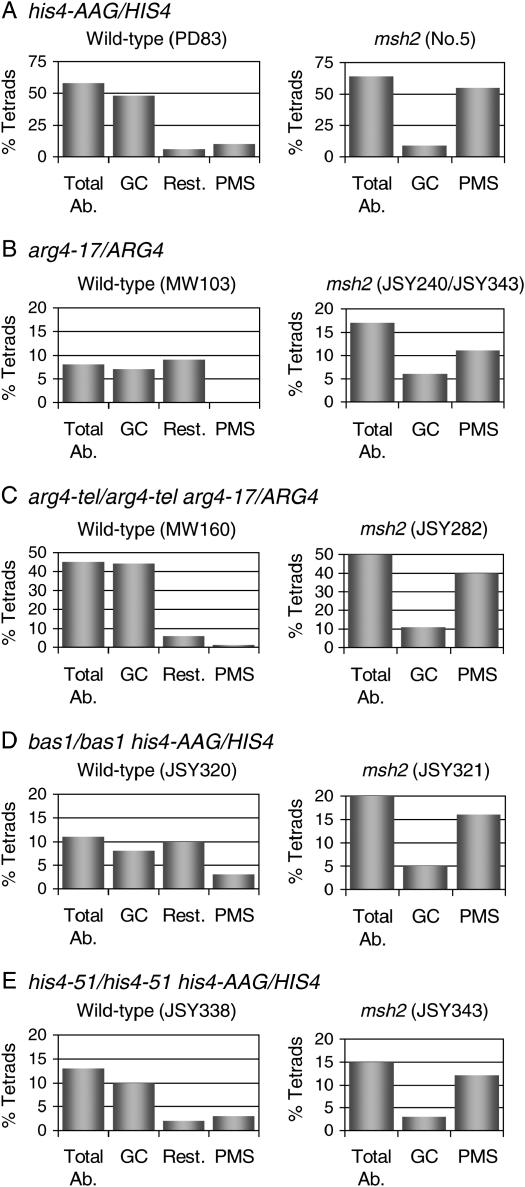
Figure 1.—
Comparison of segregation patterns in strains with varying levels of DSB hotspot activity. The observed percentages of total aberrant segregation, gene conversion, and postmeiotic segregation events are indicated as Total Ab., GC, and PMS, respectively. As described in the results, we calculated restoration events (Rest.) by comparing the frequencies of aberrant segregation in wild-type and msh2 strains. (A) Strains with wild-type DSB frequency at the HIS4 hotspot. (B) Strains with wild-type DSB frequencies at the ARG4 and DED81 hotspots. (C) Strains homozygous for an insertion of telomeric sequence upstream of ARG4 (arg4-tel) that increases the DSB frequency at the ARG4 hotspot and decreases the DSB frequency at the DED81 hotspot. (D) Strains homozygous for the bas1 deletion, a deletion that eliminates the DSB located immediately upstream of HIS4. (E) Strains homozygous for the his4-51 mutation. This mutation eliminates the Rap1p-binding site upstream of HIS4 and eliminates the HIS4-associated DSB.
One difference between the HIS4 and ARG4 loci is the strength of the nearby double-strand breaks that initiate recombination. In our strain background, the DSB site located upstream of HIS4 is one of the strongest hotspots in the genome (ranked second of all 6000 ORFs), whereas the ARG4 recombination hotspot is ranked 134th (Gerton et al. 2000). To determine whether the frequency of a local DSB would affect the conversion/restoration ratio, we examined strains in which the rate of DSB formation upstream of ARG4 was elevated and other strains in which the strength of DSB formation upstream of HIS4 was decreased.
The strength of the ARG4 hotspot affects the conversion-type/restoration-type repair ratio:
We previously showed (White et al. 1993; Fan et al. 1995) that a short (63 bp) insertion of telomeric DNA located upstream of the ARG4 gene (arg4-tel) significantly elevated the frequency of aberrant segregation and the frequency of DSB formation at the ARG4 hotspot; the position of the DSB in the arg4-tel strain, however, is approximately the same as the DSB in the wild-type strain (Fan et al. 1995). We examined the frequency of aberrant segregation of the arg4-17 mismatch in strains homozygous for the arg4-tel insertion that are either proficient or deficient (msh2) for MMR. As expected from our previous study, the frequency of aberrant segregation was very significantly elevated by the arg4-tel insertion, from ∼8 to ∼50%. In addition, the repair of the mismatch involving the arg4-17 allele was strongly biased toward conversion (Figure 1C). Thus, by increasing local DSB formation at the ARG4 hotspot, we were able to alter the ratio of conversion-type to restoration-type MMR to one that is similar to that observed at HIS4.
Effects of reducing HIS4 hotspot activity on the ratio of conversion- to restoration-type repair:
White et al. (1993) previously showed that the activity of the HIS4 recombination hotspot required the binding of the transcription factors Bas1p, Bas2p, and Rap1p. When the gene encoding Bas1p is deleted, aberrant segregation at HIS4 is significantly reduced, but not completely eliminated. A similar effect is observed when the Rap1p-binding site upstream of HIS4 is disrupted by the his4-51 mutation (White et al. 1991). We constructed isogenic diploids homozygous for the bas1 deletion or the his4-51 mutation, heterozygous for his4-AAG and arg4-17, and proficient or deficient (as a consequence of the msh2 mutation) for MMR. We found that the bas1 deletion (strain JSY320) and the his4-51 mutation (strain JSY338) reduced aberrant segregation of his4-AAG by ∼75% (Table 3) without affecting the frequency of aberrant segregation of arg4-17 (Table 4). On the basis of our observations of the ARG4 locus described above, we expected that reduction in the activity of the HIS4 hotspot would reduce conversion-type repair and increase restoration-type repair. Data from the bas1 strains (JSY320 and JSY321) were consistent with this expectation (Table 3 and Figure 1D) with conversion-type repair and restoration-type repair representing 8 and 10% of the tetrads, respectively.
The data from the his4-51 strains (JSY338 and JSY343), however, were different from those of the bas1 strains (Table 3 and Figure 1E). The comparison between the his4-51 and his4-51 msh2 strains indicates that there was little restoration-type repair in the his4-51 strain; conversion-type repair and restoration-type repair represent 10 and 2% of the his4-51 tetrads, respectively (Table 3 and Figure 1E). The difference in the results obtained with the bas1 strains and the his4-51 strains will be discussed in detail below.
DISCUSSION
Most of our understanding of the MMR machinery is based on studies of the correction of misincorporation errors of DNA polymerase (the spellchecker function). In this study, we examine the effects of the known MutS and MutL homologs, as well various nucleases implicated in MMR, on the repair of mismatches (base–base or a four-base loop) formed during meiotic recombination. In addition to confirming the importance of the Msh2/Msh6/Mlh1/Pms1 complex in the repair of base–base mismatches and the Msh2/Msh3/Mlh1/Pms1 complex in the repair of a four-base loop, we demonstrated the existence of a loop-repair complex that is Mlh1p independent but dependent on Mlh3p. We showed that the nuclease activities of Exo1p, Rad1p, Rad27p, and DNA polymerase δ have no significant role in meiotic MMR unless their functions are redundant. We also found that the ratio of conversion-type repair relative to restoration-type repair can be altered by changing the level of local DSBs.
Repair of a base–base mismatch (A/A, T/T) and a four-base loop:
In agreement with the conclusions of others (reviewed by Surtees et al. 2004), most base–base repair was equally dependent on Msh2p, Msh6p, Mlh1p, and Pms1p, as expected if these proteins function in MMR together (Figure 2). None of the nuclease-encoding genes that we tested had a significant effect on the efficiency of mismatch repair. Since it is clear the Exo1p has a role in the excision step of mismatches generated by DNA polymerase misincorporation, our results indicate either that the role of Exo1p is functionally redundant with some other nuclease during meiotic MMR or that meiosis-specific MMR is accomplished using an untested nuclease.
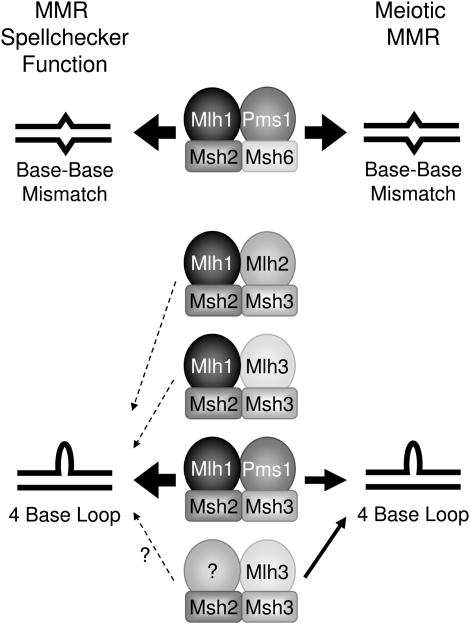
Figure 2.—
Comparison of the MutL and MutS homologs involved in the MMR spellchecker function (repair of errors introduced by DNA polymerase) and the MMR of mismatches formed during meiotic recombination. The relative contributions of each complex to the repair of base–base mismatches and four-base loop substrates are indicated by the arrow size. Msh2p/Msh6p/Mlh1p/Pms1p is involved in MMR of base–base mismatches in both contexts. Msh2p/Msh3p/Mlh1p/Pms1p initiates the majority of repair of four-base loops for the spellchecker function, with Msh2p/Msh3p/Mlh1p/Mlh2p and Msh2p/Msh3p/Mlh1p/Mlh3p making very small contributions (Wang et al. 1999; Harfe and Jinks-Robertson 2000). The meiotic repair of four-base loops is initiated primarily by Msh2p/Msh3p/Mlh1p/Pms1p, but an Mlh1-independent, Mlh3p-dependent complex (possibly Msh2p/Msh3p/Mlh3p/Mlh3p) is responsible for about one-third of the repair events.
We found that at least two types of complexes are involved in the meiotic repair of a four-base loop. The strong effects of the msh2, msh3, pms1, and mlh1 mutations argue that most repair events involve a complex of Msh2p/Msh3p/Mlh1p/Pms1p. Since the mlh1 and pms1 mutations have significantly less effect than msh2, however, some MMR is Msh2p dependent, but Mlh1p independent. Since the mlh3 mutation has a significant effect on the efficiency of repair of the four-base loop, one obvious candidate for such a complex involves a dimer of Mlh3p, either a heterodimer with a MutL homolog (other than Mlh1p) or a homodimer (Figure 2).
In a previous study, based on two-hybrid and co-immunoprecipitation analysis, Wang et al. (1999) argued that all MutL heterodimers in yeast contained Mlh1p. One explanation of this discrepancy with our data is that the stability of the MutL complexes or the levels of the MutL proteins may be different in vegetative and meiotic cells. Indeed, microarray data indicate that 2–5 hr after induction of sporulation the expression levels of MLH3, PMS1, and MSH2 are elevated twofold over vegetative expression levels, while levels of MLH1, MLH2, MSH3, and MSH6 remain constant (Chu et al. 1998). A second possible explanation is that the stability of the dimers formed between MutL homologs is affected by their interaction with the MutS homologs, an effect that would not be seen in the physical studies. Finally, we cannot exclude the possibility that certain hetero- or homodimers involving the MutL homologs are found in mlh1 strains, but not in wild-type strains.
Previously, Wang et al. (1999) found no effect of the mlh3 mutation on the efficiency of repair of a four-base loop. Since this issue was examined in a strain with very low levels of aberrant segregation (1.3%) and since the effect of the mlh3 mutation is smaller than that of mlh1 or pms1, these data are not in significant disagreement with ours. In addition, Harfe and Jinks-Robertson (2000) showed that the mlh3 mutation decreased the stability of a mononucleotide tract (10 guanine bases) ∼20-fold relative to wild type, whereas the msh2 mutation decreased the tract stability ∼10,000-fold. This result argues that Mlh3p has a minor role in the correction of a one-base loop, the substrate expected from DNA polymerase slippage of a mononucleotide tract. On the basis of our results, we predict that the mlh3 mutation might have a substantially stronger effect on the mitotic stability of a microsatellite in which the repeating unit is 4 bp.
Residual gene conversion in MMR-deficient strains:
It has been clear since the discovery of mutations that affect MMR (Williamson et al. 1985) that none of these mutations eliminates gene conversion, demonstrating that some mismatches are corrected independently of the canonical MMR system. There is evidence for a short-patch repair system in S. cerevisiae (Coic et al. 2000; E. Hoffman, J. Meadows and R. Borts, personal communication) that is independent of the nucleotide excision repair proteins, although no other proteins involved in this repair process have been identified. Alternatively, some fraction of gene conversion events may occur through a process that does not involve MMR. Merker et al. (2003) found that a small fraction (∼5%) of conversion events at the HIS4 locus appeared to reflect break-induced replication (BIR). In BIR events, a broken end from one chromosome invades and replicates another chromosome, resulting in a gene conversion event for a large chromosomal region that does not involve MMR.
Context-specific efficiency of MMR:
We noted previously that the same base–base mismatch is repaired with nearly 100% efficiency at the ARG4 locus, but is repaired with only 80–90% efficiency at the HIS4 locus (Welz-Voegele et al. 2002). This result is confirmed in this study. This difference may reflect an effect of the local sequence context on the repair of a mismatch. Alternatively, different chromosomal regions may have different efficiencies of DNA mismatch repair. An argument in favor of the latter possibility is that the rate of microsatellite alterations of the same reporter gene placed in a number of genomic locations varies by >10-fold in a MMR-proficient strain, but varies only 2-fold in a MMR-deficient strain (Hawk et al. 2005).
Genetic regulation of the rate of crossovers and aberrant segregation:
Several mutations (msh4, msh5, mlh1, mlh3, exo1, and his4-51) reduced the frequency of crossovers for one or more intervals on chromosome III. From previous studies, mlh1, mlh3, msh4, msh5, and exo1 were expected to reduce crossovers (reviewed by Hoffmann and Borts 2004), although the mechanistic roles of these proteins in crossing over is not yet clear. As already discussed in the results, we found that msh4, msh5, exo1, and pol3-01 also decreased aberrant segregation of the his4-AAG allele (Table 3). There was also a significant decrease in the aberrant segregation frequency in the mlh2 strain. This effect of the mlh2 mutation may be strain specific, since Abdullah et al. (2004) reported that mlh2 mutants had elevated levels of aberrant segregation at a variety of loci, including HIS4. Alternatively, the effect of the mlh2 mutation may reflect the distance between the initiating DSB and the mismatch. In the Abdullah et al. (2004) study, elevated levels of aberrant segregation were observed only for markers in HIS4 located >500 bp from the initiating lesion.
The conversion- to restoration-type repair ratio:
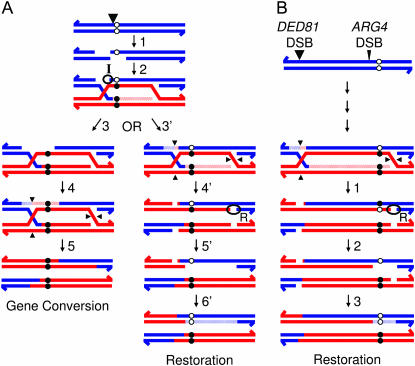
The meiotic repair of a mismatch either can lead to a gene conversion event or can restore Mendelian segregation. Although restoration events are difficult to measure directly, the frequency of such events can be estimated by subtracting the frequency of aberrant segregation in a MMR-proficient strain from the frequency of aberrant segregation in a MMR-deficient strain (details discussed in the results). In confirmation of our previous results, we found that the base–base mismatch represented by the his4-AAG allele was primarily corrected by conversion-type repair (Figure 1A). One explanation of such a bias is that the nick resulting from the DSB that initiates recombination [which we will call the I (Initiating) nick] usually directs the excision of mismatches, if the mismatch is located near the DSB site (Porter et al. 1993); this repair would yield exclusively conversion-type repair (Figure 3A, steps 3–5). This type of repair has also been termed “early repair” (Foss et al. 1999).
Figure 3.—
Nick-directed mismatch repair. (A) Mismatch repair directed by the nick that initiates DSB formation (Detloff et al. 1992; Porter et al. 1993) or by the nick that is involved in resolving the recombination intermediate (Foss et al. 1999; “late repair”). Recombination is initiated by a DSB (indicated by large arrowhead) on the wild-type (blue) chromosome, and the broken ends are resected (step 1). After strand invasion, DNA synthesis extends the D-loop, resulting in a mismatch within the heteroduplex DNA (step 2). Steps 3–5 illustrate repair that is directed by the nick remaining at the initiation site (the I nick). In step 3, DNA from the I nick through the mismatch is excised. DNA synthesis subsequently fills in the gap, duplicating the mutant information (step 4). I-nick-directed repair results in a gene conversion event (step 5). Steps 3′–6′ depict repair that is directed by nicks involved in resolving the Holliday junctions (the R nicks). In steps 3′ and 4′, the DNA strands that have been involved in the exchange are nicked (shown by small arrowheads). In step 5′, DNA is excised from the R nick through the mismatch. DNA synthesis then replicates the wild-type information (step 6′), resulting in a restoration event. (B) Repair directed by DSBs initiated in a neighboring gene. Southern analysis demonstrates that there are two preferred sites for DSB formation near ARG4, one immediately upstream of ARG4 and a second upstream of the neighboring DED81 gene (Nicolas et al. 1989; Fan et al. 1995). In wild-type strains, more DSBs are generated at DED81 than at ARG4, as indicated by the size of the arrows. We suggest that the majority of restoration-type repair of the arg4-17 mismatch are the result of the processing of an R nick of a heteroduplex initiated by the DED81 DSB. A second source of restoration events is excision from an R nick of a heteroduplex initiated by the ARG4 DSB, as shown in A, steps 3′–6′.
Mismatches located in the HIS4 gene further from the initiating lesion have both restoration-type repair and conversion-type repair (Detloff et al. 1992). Foss et al. (1999) suggested that restoration-type repair (termed “late repair”) could be directed by the nick resulting from resolution of the Holliday junction (HJ). We label this nick “R” (Resolution) in Figure 3A (steps 3′–6′). However, if the R nick is not close enough to the mismatch to direct the excision event and/or repair has not occurred by the time the DNA is ligated following HJ resolution, the repair event could be undirected (50% conversion-type and 50% restoration-type repair). Thus, the ratio of conversion- to restoration-type repair is likely to be related to the distance of the mismatch from the two types of nicks that can direct the excision event. Since the his4-AAG-associated mismatch is located near the DNA lesion that initiates the heteroduplex, almost all repair would be directed toward conversion (Porter et al. 1993).
Unlike the mismatch involving his4-AAG, the mismatch related to the arg4-17 substitution had approximately equal frequencies of conversion-type and restoration-type MMR (Figure 1B). In the context of the model shown in Figure 3A, this result is consistent with the arg4-17-associated mismatch being equidistant from I and R nicks. This interpretation is inconsistent, however, with the observation that the distance between his4-AAG and the recombination-initiating DSB (250 bp; Fan et al. 1995; Xu and Petes 1996) is approximately the same as the distance between arg4-17 and its recombination-initiating DSB (∼330 bp; de Massy and Nicolas 1993). Our alternative interpretation is shown in Figure 3B. We argue that the heteroduplexes that produce mismatches involving the arg4-17 substitution are initiated at two different sites. Events initiated at the ARG4 hotspot result in an I nick near the mismatch and, consequently, would usually result in conversion-type repair. We suggest that heteroduplexes initiated as a strong DSB site located upstream of the neighboring gene (DED81) also contribute to mismatches involving the arg4-17 allele (Figure 3B). For this class of events, we suggest that the repair of the mismatch is usually directed by an R nick, leading to restoration. Alternatively, the mismatch initiated at DED81 might be corrected by a mechanism that is not nick directed and yields an equal frequency of conversion-type and restoration-type repair.
The hypothesis that the segregation frequency of the arg4-17 marker is affected by DSBs initiated at more than one site is supported by a number of arguments. First, in our genetic background, the DSB site located upstream of DED81 is considerably stronger than that located upstream of ARG4 (Fan et al. 1995). Second, in the same genetic background, heteroduplexes initiated upstream of HIS4 often extend to distances >2.5 kb (Detloff et al. 1992; Merker et al. 2003), a distance less than that between the DED81-associated DSB and the arg4-17 substitution. Third, recombination events at the HIS4 locus are initiated at multiple DSB sites (Merker et al. 2003). Fourth, the model shown in Figure 3B is consistent with our observation that increasing the strength of the ARG4-associated DSB (arg4-tel insertion) results in an increase in conversion-type repair relative to restoration-type repair (Figure 1C).
This model also predicts that reduction in the strength of the HIS4-associated DSB might alter the ratio of conversion-type to restoration-type repair, since the frequency of repair events directed by the I nick would be reduced. The HIS4-associated DSB hotspot requires the binding of the transcription factors Rap1p, Bas1p, and Bas2p, and strains with either a bas1 deletion or a mutated Rap1p-binding site (his4-51) eliminate the DSB located upstream of HIS4 (White et al. 1993; Fan et al. 1995). In the wild-type strain, the calculated ratio of conversion-type repair to restoration-type repair is 8:1; this ratio is reduced to ∼1:1 by the bas1 deletion (Figure 1D). In contrast, in the strain with the his4-51 mutation, the calculated ratio of conversion-type to restoration-type repair was not significantly reduced.
Why does the bas1 deletion affect the ratio of conversion-type to restoration-type repair differently from the his4-51 mutation? As discussed above, in a MMR-deficient strain, the frequency of aberrant segregation is primarily a reflection of the frequency of heteroduplex formation at the position of the heterozygous marker, whereas, in a MMR-proficient strain, the frequency of aberrant segregation reflects the frequency of heteroduplex formation and the ratio of conversion-type to restoration-type repair. Since both the bas1 and the his4-51 strains lack the DSB located upstream of HIS4 that is responsible for most recombination at this locus (Fan et al. 1995), the heteroduplexes that result in aberrant segregation of his4-AAG are a consequence of DSBs located at other sites (as shown previously in Merker et al. 2003). We suggest that the rate of DSB formation at these other sites and/or the distribution of I and R nicks are different in bas1 and his4-51 strains. Although this suggestion is somewhat ad hoc, the effects of Bas1p and Rap1 on local chromatin structure at the HIS4 locus are likely to be different, since Rap1p binding is required for binding of Bas1p upstream of HIS4, but Bas1p binding is not required for the binding of Rap1p (Morse 2000). Given these uncertainties, the experiment in which we altered the DSB frequency upstream of ARG4 is the more definitive study.
Summary:
We found that a complex of Msh2p/Msh6p/Mlh1p/Pms1p is required for repair of base–base mismatches generated during meiotic recombination. At least two complexes initiate repair of the four-base loop: Msh2p/Msh3p/Mlh1p/Pms1p is responsible for about two-thirds of MMR and the remaining repair events are initiated by an Mlh1p-independent, Mlh3p-dependant complex. In addition, our results indicate that the rate of aberrant segregation for a single marker and the direction in which a mismatch is repaired are often influenced by DSBs located at several positions. This conclusion may be relevant to the variability in the effects of mutations catalyzing MMR observed by different researchers, working in different genetic backgrounds and analyzing different loci.
Acknowledgments
We thank M. Gawel for assistance with tetrad dissection and D. Jenness for the gift of plasmid pDJ173. We are grateful to J. L. Argueso and F. J. Lemoine for helpful discussions regarding the manuscript and to P. Luedi for advice on the statistical analysis. This work was supported by National Institutes of Health grant GM24110.
References
- Abdullah, M. F., E. R. Hoffmann, V. E. Cotton and R. H. Borts, 2004. A role for the MutL homologue MLH2 in controlling heteroduplex formation and in regulating between two different crossover pathways in budding yeast. Cytogenet. Genome Res. 107: 180–190. [DOI] [PubMed] [Google Scholar]
- Alani, E., R. A. Reenan and R. D. Kolodner, 1994. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 137: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso, J. L., J. Wanat, Z. Gemici and E. Alani, 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168: 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57: 289–300. [Google Scholar]
- Börner, G. V., N. Kleckner and N. Hunter, 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein et al., 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- Coic, E., L. Gluck and F. Fabre, 2000. Evidence for short-patch mismatch repair in Saccharomyces cerevisiae. EMBO J. 19: 3408–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin, N., L. Dzantiev, F. A. Karyov and P. Modrich, 2005. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 280: 39752–39761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy, B., and A. Nicolas, 1993. The control in cis of the position and the amount of the ARG4 meiotic double-strand break of Saccharomyces cerevisiae. EMBO J. 12: 1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff, P., J. Sieber and T. D. Petes, 1991. Repair of specific base pair mismatches formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff, P., M. A. White and T. D. Petes, 1992. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics 132: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzantiev, L., N. Constantin, J. Genschel, R. R. Iyer, P. M. Burgers et al., 2004. A defined human system that supports bidirectional mismatch-provoked excision. Mol. Cell 15: 31–41. [DOI] [PubMed] [Google Scholar]
- Fan, Q., F. Xu and T. D. Petes, 1995. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15: 1679–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss, H. M., K. J. Hillers and F. W. Stahl, 1999. The conversion gradient at HIS4 of Saccharomyces cerevisiae. II. A role for mismatch repair directed by biased resolution of the recombinational intermediate. Genetics 153: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschel, J., L. R. Bazemore and P. Modrich, 2002. Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 277: 13302–13311. [DOI] [PubMed] [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34: 359–399. [DOI] [PubMed] [Google Scholar]
- Harfe, B. D., B. K. Minesinger and S. Jinks-Robertson, 2000. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol. 10: 145–148. [DOI] [PubMed] [Google Scholar]
- Hawk, J. D., L. Stefanovic, J. C. Boyer, T. D. Petes and R. A. Farber, 2005. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc. Natl. Acad. Sci. USA 102: 8639–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, K. A., and S. Keeney, 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 101: 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz, I., and R. E. Jensen, 1991. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 194: 132–146. [DOI] [PubMed] [Google Scholar]
- Hoffmann, E. R., and R. H. Borts, 2004. Meiotic recombination intermediates and mismatch repair proteins. Cytogenet. Genome Res. 107: 232–248. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, N. M., L. Ponte and C. Hasley, 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9: 1728–1739. [DOI] [PubMed] [Google Scholar]
- Hunter, N., and R. H. Borts, 1997. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11: 1573–1582. [DOI] [PubMed] [Google Scholar]
- Kearney, H. M., D. T. Kirkpatrick, J. L. Gerton and T. D. Petes, 2001. Meiotic recombination involving heterozygous large insertions in Saccharomyces cerevisiae: formation and repair of large, unpaired DNA loops. Genetics 158: 1457–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazanehdari, K. A., and R. H. Borts, 2000. EXO1 and MSH4 differentially affect crossing-over and segregation. Chromosoma 109: 94–102. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, D. T., and T. D. Petes, 1997. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature 387: 929–931. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, D. T., J. R. Ferguson, T. D. Petes and L. S. Symington, 2000. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics 156: 1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska, R. J., L. Stefanovic, H. T. Tran, M. A. Resnick, D. A. Gordenin et al., 1998. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol. Cell. Biol. 18: 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., T. Kane, C. Tipper, P. Spatrick and D. D. Jenness, 1999. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol. Cell. Biol. 19: 3588–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel, L., J. Bhargava and G. S. Roeder, 2004. A role for DNA polymerase δ in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae. Genetics 167: 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, T. M., and O. Fleck, 2004. DNA repair nucleases. Cell. Mol. Life Sci. 61: 336–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker, J. D., M. Dominska and T. D. Petes, 2003. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 165: 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich, P., and R. Lahue, 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65: 101–133. [DOI] [PubMed] [Google Scholar]
- Morrison, A., A. L. Johnson, L. H. Johnston and A. Sugino, 1993. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 12: 1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, R. H., 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16: 51–53. [DOI] [PubMed] [Google Scholar]
- Nag, D. K., M. A. White and T. D. Petes, 1989. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature 340: 318–320. [DOI] [PubMed] [Google Scholar]
- Nicolas, A., D. Treco, N. P. Schultes and J. W. Szostak, 1989. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 338: 35–39. [DOI] [PubMed] [Google Scholar]
- Perkins, D. D., 1949. Biochemical mutants in the smut fungus Ustilago maydis. Genetics 34: 607–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., and D. Botstein, 1977. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc. Natl. Acad. Sci. USA 74: 5091–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., R. E. Malone and L. Symington, 1991. Recombination in yeast, pp. 407–521 in The Molecular and Cellular Biology of the Yeast Saccharomyces, edited by J. Broach, E. W. Jones and J. R. Pringle. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Porter, S. E., M. A. White and T. D. Petes, 1993. Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics 134: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan, R. A., and R. D. Kolodner, 1992. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics 132: 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald, P., and G. S. Roeder, 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1982. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sia, E. A., and D. T. Kirkpatrick, 2005. The yeast MSH1 gene is not involved in DNA repair or recombination during meiosis. DNA Rep. 4: 253–261. [DOI] [PubMed] [Google Scholar]
- Stapleton, A., and T. D. Petes, 1991. The Tn3 β-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics 127: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic, L., R. Brun and J. Jiricny, 2004. Mismatch repair and DNA damage signaling. DNA Rep. 3: 1091–1101. [DOI] [PubMed] [Google Scholar]
- Sugawara, N., F. Paques, M. Colaiacovo and J. E. Haber, 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94: 9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees, J. A., J. L. Argueso and E. Alani, 2004. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet. Genome Res. 107: 146–159. [DOI] [PubMed] [Google Scholar]
- Tishkoff, D. X., A. L. Boerger, P. Bertrand, N. Filosi, G. M. Gaida et al., 1997. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. USA 94: 7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, P. T., J. A. Simon and R. M. Liskay, 2001. Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98: 9760–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi, H., and H. Ogawa, 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11: 2221–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar, A., A. B. Buermeyer, J. A. Simon, D. C. Thomas, A. B. Clark et al., 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87: 65–73. [DOI] [PubMed] [Google Scholar]
- Vedel, M., and A. Nicolas, 1999. CYS3, a hotspot of meiotic recombination in Saccharomyces cerevisiae. Effects of heterozygosity and mismatch repair functions on gene conversion and recombination intermediates. Genetics 151: 1245–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wang, T. F., N. Kleckner and N. Hunter, 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96: 13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz-Voegele, C., J. E. Stone, P. T. Tran, H. M. Kearney, R. M. Liskay et al., 2002. Alleles of the yeast Pms1 mismatch-repair gene that differentially affect recombination- and replication-related processes. Genetics 162: 1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. H., K. Lusnak and S. Fogel, 1985. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature 315: 350–352. [DOI] [PubMed] [Google Scholar]
- White, M. A., M. Wierdl, P. Detloff and T. D. Petes, 1991. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc. Natl. Acad. Sci. USA 88: 9755–9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, M. A., M. Dominska and T. D. Petes, 1993. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90: 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, M. S., J. C. Game and S. Fogel, 1985. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1–1 and pms1–2. Genetics 110: 609–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F., and T. D. Petes, 1996. Fine-structure mapping of meiosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics 143: 1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]