TABLE 2.
Descriptive statistics and estimates of nucleotide sequence diversity for a common set of 25 samples at 18 loci in wild barley
| Gene | n | h | Aligned length, bp | Sp |
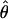 W × 10−3 W × 10−3
|
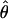 π ×10−3 π ×10−3
|
T | Wall's B | Rm | Rh | Rs | Rl | Ru |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H. vulgare ssp. spontaneum | |||||||||||||
| Adh1 | 25 | 11 | 1362 | 6 | 2.73 (±1.11) | 2.07 | −0.926 | 0.154 | 0 | 0 | 0 | 0 | 0 |
| Adh2 | 25 | 19 | 1980 | 14 | 4.84 (±1.72) | 3.19 | −1.289 | 0.057 | 2 | 2 | 2 | 2 | 2 |
| Adh3 | 25 | 21 | 1873 | 81 | 15.42 (±5.11) | 22.42 | 1.734 | 0.423 | 2 | 2 | 5 | 5 | 6 |
| α-amy1 | 25 | 5 | 856 | 3 | 3.10 (±1.36) | 1.27 | −1.948 | 0.222 | 0 | 0 | 0 | 0 | 0 |
| Cbf3 | 28 | 10 | 1514 | 22 | 4.52 (±1.64) | 4.43 | −0.071 | 0.160 | 2 | 3 | 3 | 3 | 3 |
| Dhn1 | 24 | 16 | 1538 | 37 | 18.70 (±6.36) | 13.18 | −1.161 | 0.056 | 7 | 11 | 11 | 11 | 17 |
| Dhn4 | 24 | 12 | 1072 | 31 | 14.13 (±4.97) | 17.18 | 0.831 | 0.381 | 5 | 6 | 6 | 6 | 8 |
| Dhn5 | 24 | 19 | 1088 | 25 | 11.70 (±4.09) | 10.81 | −0.295 | 0.239 | 3 | 7 | 7 | 7 | 8 |
| Dhn7 | 28 | 19 | 1389 | 50 | 16.72 (±6.21) | 14.01 | −0.622 | 0.185 | 6 | 11 | 11 | 13 | 16 |
| Dhn9 | 25 | 12 | 1011 | 9 | 4.90 (±1.91) | 3.91 | −0.725 | 0.118 | 1 | 1 | 1 | 1 | 1 |
| Faldh | 25 | 11 | 1091 | 17 | 5.67 (±2.12) | 5.71 | −0.405 | 0.333 | 0 | 0 | 0 | 1 | 1 |
| G3pdh | 26 | 13 | 2010 | 45 | 7.93 (±2.64) | 9.90 | 0.823 | 0.536 | 1 | 1 | 1 | 3 | 3 |
| ORF1 | 27 | 17 | 1533 | 22 | 6.16 (±2.16) | 5.18 | −1.129 | 0.196 | 1 | 1 | 1 | 1 | 1 |
| 5′Pepc | 25 | 6 | 2019 | 1 | 0.66 (±0.35) | 0.23 | −1.841 | — | 0 | 0 | 0 | 0 | 0 |
| Pepc | 25 | 8 | 1154 | 3 | 1.15 (±0.61) | 1.14 | −0.023 | — | 0 | 0 | 0 | 0 | 0 |
| Stk | 26 | 15 | 1057 | 20 | 9.29 (±3.27) | 6.77 | −1.019 | 0.111 | 1 | 3 | 3 | 3 | 3 |
| Vrn1 | 19 | 12 | 1262 | 10 | 3.79 (±1.48) | 3.57 | −0.216 | 0.077 | 2 | 2 | 3 | 2 | 3 |
| Waxy | 28 | 22 | 1232 | 25 | 9.12 (±3.12) | 7.86 | −0.521 | 0.238 | 6 | 12 | 12 | 12 | 16 |
| External H. vulgare ssp. spontaneum data | |||||||||||||
| GSP | 33 | 25 | 1802 | 38 | 19.43 (±5.89) | 7.52 | −2.326 | 0.164 | 9 | 9 | 13 | 10 | 13 |
| hina | 33 | 22 | 1475 | 44 | 10.87 (±3.45) | 10.00 | −0.295 | 0.067 | 9 | 10 | 13 | 9 | 13 |
| hinb | 33 | 26 | 3373 | 134 | 14.08 (±4.29) | 9.16 | −1.331 | 0.080 | 6 | 7 | 10 | 7 | 8 |
| D. melanogaster | |||||||||||||
| bagpipe | 27 | 12 | 1402 | 27 | 6.16 (±2.12) | 7.23 | 0.647 | 0.300 | 6 | 7 | 9 | 9 | 10 |
| CG3588 | 44 | 26 | 1332 | 34 | 11.64 (±3.45) | 8.23 | −1.043 | 0.050 | 13 | 13 | 30 | 31 | — |
| Est6 | 50 | 22 | 2332 | 77 | 13.00 (±3.77) | 11.30 | −0.463 | 0.178 | 15 | 15 | 20 | 21 | 31 |
| Idgf1 | 20 | 18 | 1958 | 72 | 11.61 (±4.03) | 14.66 | 1.073 | 0.132 | 15 | 15 | 20 | 23 | 35 |
| Idgf3 | 20 | 17 | 2401 | 48 | 8.33 (±2.95) | 8.76 | 0.209 | 0.275 | 11 | 11 | 16 | 16 | 21 |
| Notch5′ | 50 | 29 | 1480 | 24 | 8.38 (±2.55) | 4.51 | −1.598 | 0.096 | 9 | 13 | 27 | 24 | — |
| polehole | 22 | 18 | 2259 | 40 | 6.69 (±2.31) | 6.69 | −0.001 | 0.212 | 13 | 13 | 20 | 20 | 31 |
| tinman | 29 | 16 | 2428 | 26 | 4.29 (±1.47) | 5.15 | 0.734 | 0.316 | 2 | 2 | 2 | 2 | 2 |
| vermilion | 71 | 36 | 2081 | 68 | 11.35 (±3.00) | 8.84 | −0.757 | 0.047 | 27 | 27 | 60 | 56 | — |
| yEst6 | 22 | 12 | 2332 | 72 | 10.64 (±3.64) | 11.97 | 0.502 | 0.438 | 4 | 4 | 4 | 4 | 6 |
| D. pseudoobscura | |||||||||||||
| Adh | 139 | 118 | 4736 | 217 | 2.66 (±6.14) | 11.72 | −1.845 | 0.0424 | 54 | 61 | 155 | — | — |
| D. simulans | |||||||||||||
| Notch 3′ | 22 | 16 | 1578 | 26 | 5.91 (±2.16) | 6.92 | 0.664 | 0.091 | 9 | 9 | 17 | 17 | 24 |
| Notch 5′ | 22 | 20 | 1411 | 28 | 7.71 (±2.79) | 8.03 | 0.162 | 0.054 | 10 | 10 | 19 | 19 | 27 |
| Z. mays | |||||||||||||
| Adh | 25 | 11 | 1435 | 40 | 11.93 (±4.07) | 12.58 | 0.210 | 0.138 | 8 | 8 | 9 | 9 | 11 |
| Glb1 | 23 | 20 | 1196 | 50 | 25.19 (±8.36) | 19.87 | −0.843 | 0.082 | 14 | 14 | 21 | 21 | 32 |
| Umc128 | 23 | 15 | 1011 | 23 | 15.45 (±5.68) | 19.43 | 0.979 | 0.207 | 5 | 5 | 7 | 7 | 9 |
| Umc230 | 22 | 12 | 1243 | 17 | 17.79 (±6.56) | 12.79 | −1.082 | 0.0333 | 3 | 3 | 4 | 4 | 5 |
| Zfl2 | 29 | 28 | 4205 | 82 | 16.73 (±5.22) | 10.53 | −1.439 | 0.115 | 24 | 26 | 50 | 44 | — |
See Table 1 for symbols used. For 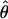 W, standard deviation is shown, based on no recombination.
W, standard deviation is shown, based on no recombination.
