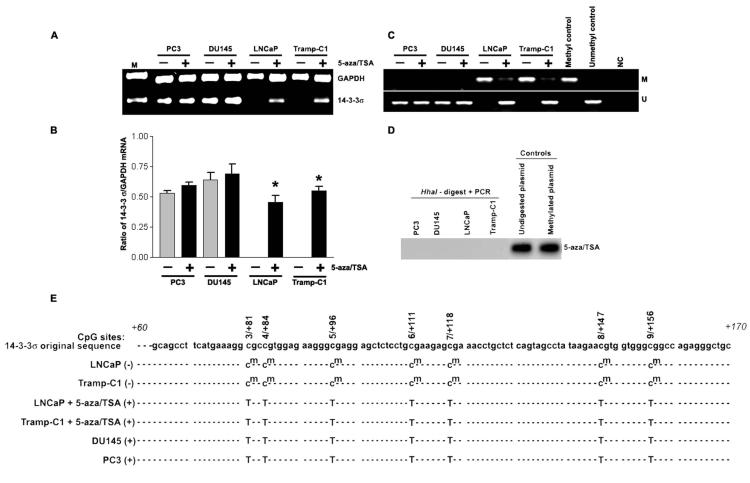
Figure 3.
- mRNA expression of 14-3-3σ was analyzed using reverse transcription-PCR in LNCaP, DU145, PC3 and Tramp-C1 prostate cancer cells treated with either DMSO (control) or 10 μM 5-aza followed by 50 nM TSA, as described in Materials and Methods. GAPDH mRNA was co-amplified as a loading control and expression standard.
- Bar diagram showing densitometry quantified data of 14-3-3σ/GAPDH mRNA ratios from three independent experiments. Each bar represents triplicate analyses of mean ± SD where significant difference from cells treated with vehicle alone (control) is represented by an asterisk * (P <0.05).
- MSP assay on DNA isolated from 5-aza/TSA-treated or untreated cell lines PC3, DU145, LNCaP and Tramp-C1. Methylation-specific primers generated the PCR products labeled with “M”. Those labeled with “U” were generated by primers specific for unmethylated DNA. Positive and negative controls were described in Materials and Methods.
- HhaI-based DNA methylation assay on DNA isolated from 5-aza and TSA-treated PC3, DU145, LNCaP and Tramp-C1 cells. SssI-methylated, 14-3-3σ plasmid DNA and undigested 14-3-3σ plasmid DNA were used as positive controls.
- Bisulfite sequencing analysis of 14-3-3σ promoter CpG island methylation in 5-aza/TSA treated or untreated prostate cancer cells. Seven potential CpG sites (3/+81 to 9/+156, genomic positions 8721, 8724, 8736, 8751, 8758, 8787, and 8796; Accession No. AF029081) were analyzed by methylation-specific PCR followed by cloning and DNA sequencing. Methylated cytosines remain cytosines (cm), whereas unmethylated cytosines are converted thymidine (T). Absence of 14-3-3σ expression (−) and presence of 14-3-3σ expression (+). Results are representative of at least 4 different DNA sequence analyses.