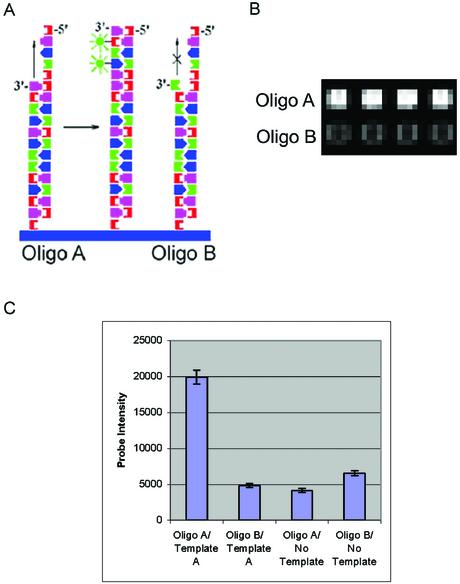
Figure 6.
Sequence-specific primer extension reactions. Arrays were synthesized with alternating rows of oligonucleotide sequences designated oligo A (5′-AGG TCA TTA CAG CGA GAG-3′) and oligo B (5′-AGG TCA TTA CAG CGA GAC-3′), which are identical except for the 3′ nucleotide. (A) Primer extension scheme showing hybridization of template A (5′-TGA CCT ATA ATC CTC TCG CTG TAA TGA CCT-3′) to the two array oligos. Klenow DNA polymerase is used to extend the 3′ end of the array oligos, and during the extension, labeled nucleotides are covalently attached to the array surface. Oligo A should extend with greater efficiency than oligo B, due to the 3′ mismatch in oligo B with template A. (B) Detail showing signal generated from oligo A and oligo B extended with template A. The array was scanned at a PMT gain of 520. (C) Intensity values resulting from primer extension of oligo A and oligo B with template A or no template.