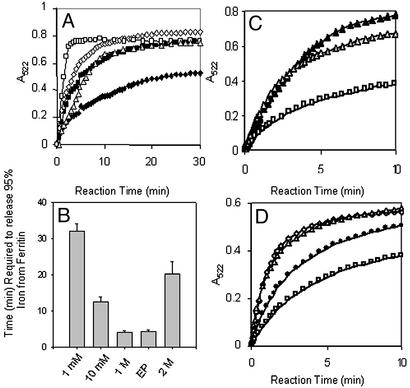
Figure 2.
Chaotropes decrease the time required to chelate Fe in the ferritin biomineral, mimicking the unfolding of ferritin pores associated with amino acid substitution of phylogenetically conserved pore residues. Recombinant ferritin protein (the frog H-WT), isolated with <5 Fe per 24 assembled subunits, was mineralized by mixing a solution of 2.08 μM protein in 0.10 M Mops/0.1 M NaCl, pH 7, with a solution of FeSO4 in 0.001 M HCl (see Experimental Methods). Reduction and chelation were triggered by mixing equal volumes of protein solution and a solution of 5 mM each of NADH/FMN/bipyridyl in 0.10 M Mops/0.1 M NaCl, pH 7 (see Experimental Methods). (A) Progress curve for formation of Fe2+-bipyridyl + urea. Chaotrope concentrations: ♦, 0; ■, 1 mM; ⋄, 10 mM; □, 1 M; ▵, 2 M. (B) Decrease in the time (minutes) required to remove all the Fe from ferritin in the presence of urea. Note that, in the absence of urea, removal of all the Fe required 150 min under these conditions (3). EP, engineered protein H-L134P. (C) Progress curves for formation of Fe2+-bipyridyl with Triton X-100. Chaotrope concentrations: □, 0; ▵, 1%; ▴, 10%. (D) Progress curve for formation of Fe2+-bipyridyl with Gdn⋅HCl. Chaotrope concentrations: □, 0; ●, 0.1 mM; ▵, 1 mM; ⋄, 10 mM.