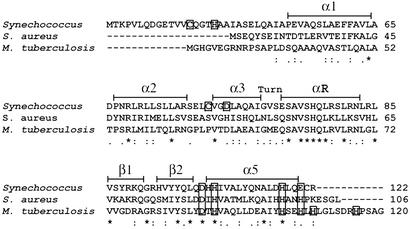
Figure 1.
A multiple sequence alignment of Synechococcus PCC 7942 SmtB (P30340), S. aureus CzrA (O85142), and M. tuberculosis NmtR (NP_218262.1) generated by CLUSTAL W (36) (Swiss-Prot/TrEMBL accession nos. for CzrA and SmtB or the GenBank accession no. for NmtR are in parentheses). Residues that are identical (*), residues that are strongly similar (:), and residues that are weakly similar (.) are denoted below the alignment. Secondary structure elements are denoted above the alignment and based on the crystal structure of apo-SmtB (34) and chemical-shift indexing for CzrA (6). Conserved metal-binding residues located in the α5 site are boxed, as are proposed ligands to the α3N site in SmtB (C14, H18, C61, D64) (17, 18).