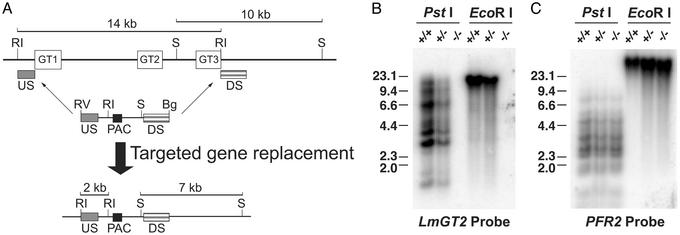
Figure 1.
Construction of the glucose transporter null mutant, Δlmgt, by targeted gene replacement. (A) Strategy for targeted gene replacement. (Upper) The LmGT gene locus including the three ORFs (open rectangles marked GT1, GT2, and GT3), the 10-kb SmaI and 14-kb EcoRI restriction fragments, and the location of the upstream (US) and downstream (DS) segments used to target homologous recombination of the gene disruption constructs. One of these disruption constructs is shown immediately below the LmGT locus and includes the US and DS segments, the ORF for the PAC selectable marker, the EcoRV and BglII terminal polylinker restriction sites, and the internal EcoRI and SmaI restriction sites. The thin arrows indicate the sites of homologous integration. (Lower) The targeted gene replacement event is indicated below the thick arrow, showing the structure of the resulting chromosomal locus and the predicted 2-kb EcoRI and 7-kb SmaI restriction fragments that are diagnostic of the correct homologous integration event. Symbols indicating restriction fragments are: RI, EcoRI; S, SmaI; RV, EcoRV; and Bg, BglII. Generation of the null mutant required a second targeted gene replacement using a similar gene disruption cassette containing a SAT marker. (B) Southern blot containing 10 μg of genomic DNA from WT parasites (+/+), heterozygous knockout line after integration of the PAC gene disruption construct (+/−), or Δlmgt null mutant (−/−) digested with PstI or with EcoRI and hybridized with a radiolabeled probe representing the LmGT2 ORF. The numbers indicate the positions and sizes (kilobase pairs) of DNA molecular weight markers. (C) The same blot shown in B after elution of the LmGT2 probe and rehybridization to the PFR2 probe that encodes an unrelated paraflagellar rod protein.