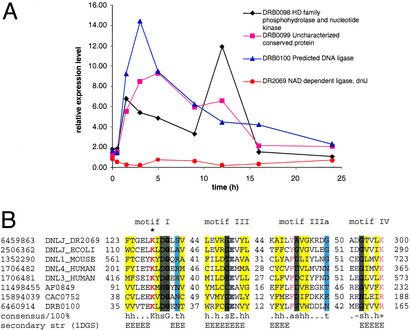
Figure 4.
Expression of a predicted operon coding for components of a potential distinct DNA repair system. (A) Expression pattern of the genes in the predicted repair operon and the NAD-dependent DNA ligase. (B) Multiple alignment of the conserved motifs common to all DNA ligases, including the predicted distinct ATP-dependent DNA ligase DRB0100 from DEIRA. The alignment was constructed manually on the basis of high-scoring sequence pairs generated by PSI-BLAST searches (47) and previously described conservation of DNA ligase motifs (32, 33). Sequences are denoted by gene identifiers and gene names from the GenBank database: NAD-dependent DNA ligase from DEIRA (DR2069), E. coli (DNLJ_ECOLI), DNA ligase IV from human (DNL4_HUMAN), DNA ligase III from human (DNL3_HUMAN), DNA ligase I from mouse (DNL3_MOUSE), predicted ATP-dependent DNA ligases from DEIRA (DRB0100), Archaeoglobus fulgidus (AF0849), and Clostridium acetobutylicum (CAC0752). The positions of the first and the last residue of the aligned region in the corresponding protein are indicated for each sequence. The numbers within the alignment represent less conserved regions that are not shown. Invariant amino acid residues are in bold type. Alignment coloring is based on the consensus shown underneath the alignment; h, hydrophobic residues (A, C, F, L, I, M, V, W, Y, T, S, and G; yellow background); t, turn-forming residues (A, C, S, T, D, E, N, V, G, and P; cyan background); s, small residues (A, C, S, T, D, V, G, and P; green background); p, positively charged residues (R and K; red); a, aromatic residues (W, Y, F, and H; magenta); −, negatively charged residues (E and D; blue). The secondary structure elements are shown according to the structure of the NAD-dependent DNA ligase from Thermus filiformis, 1DGS (48). H, extended conformation, α-helix; E, extended conformation, β-strand. The catalytic lysine is marked by an asterisk.