Abstract
We recently identified 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, the rate-limiting enzyme of the mevalonate pathway, as a potential therapeutic target of the head and neck squamous cell carcinomas (HNSCC) and cervical carcinomas (CC). The products of this complex biochemical pathway, including de novo cholesterol, are vital for a variety of key cellular functions affecting membrane integrity, cell signaling, protein synthesis, and cell cycle progression. Lovastatin, a specific inhibitor of HMG-CoA reductase, induces a pronounced apoptotic response in a specific subset of tumor types, including HNSCC and CC. The mediators of this response are not well established. Identification of differentially expressed genes represents a feasible approach to delineate these mediators as lovastatin has the potential to modulate transcription indirectly by perturbing levels of sterols and other mevalonate metabolites. Expression analysis following treatment of the HNSCC cell lines SCC9 or SCC25 with 10 µM lovastatin for 1 day showed that less than 2% (9 cDNAs) of the 588 cDNAs on this microarray were affected in both cell lines. These included diazepam-binding inhibitor/acyl-CoA-binding protein, the activated transcription factor 4 and rhoA. Because the biosynthesis of mevalonate leads to its incorporation into more than a dozen classes of end products, their role in lovastatin-induced apoptosis was also evaluated. Addition of the metabolites of all the major branches of the mevalonate pathway indicated that only the nonsterol moiety, geranylgeranyl pyrophosphate (GGPP), significantly inhibited the apoptotic effects of lovastatin in HNSCC and CC cells. Because rhoA requires GGPP for its function, this links the microarray and biochemical data and identifies rhoA as a potential mediator of the anticancer properties of lovastatin. Our data suggest that the depletion of nonsterol mevalonate metabolites, particularly GGPP, can be potential mediators of lovastatin-induced apoptosis of HNSCC and CC cells.
Keywords: HMG-CoA reductase, lovastatin, apoptosis, squamous cell carcinoma, geranylgeranyl
Introduction
Cellular processes that contribute to neoplastic transformation include deregulation of proliferation, differentiation, cell survival signaling, and apoptosis [13,15,24]. Agents that can target these pathways and induce an apoptotic or a programmed cell death response of malignant cells are potential anticancer therapeutic approaches [15,24,33]. The clinical utility of such modulators, however, has been limited due to toxicity and lack of specificity [27,33]. Refinements in target identification and validation may uncover agents with greater therapeutic potential. Using differential expression methodologies, we recently identified the enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, the rate-limiting enzyme of the mevalonate pathway, as a potential anticancer therapeutic target [7,10]. Inhibition of enzyme function elicited a potent apoptotic response in a specific subset of human cancers [7,10].
Mevalonate is a critical component of a complex biochemical pathway whose products are vital for a variety of key cellular functions including membrane integrity, cell signaling, protein synthesis, and cell cycle progression [17]. The diverse array of critical metabolic end products of this pathway, which includes de novo cholesterol, strongly suggests that physiological regulation of HMG-CoA reductase is essential for the maintenance of cellular homeostasis [17]. Lovastatin is a specific, nonreversible competitive inhibitor of HMG-CoA reductase [4,19,40], whose ability to block this critical metabolic pathway has led to its extensive clinical use as a treatment for hypercholesterolemia [4,19,40]. Based on our original observations that targeting HMG-CoA reductase induced a potent apoptotic response in neuroblastoma and acute myeloid leukemia cells, we evaluated the sensitivity of a variety of tumor cell lines to lovastatin-induced apoptosis [6,9]. Included in this survey were a variety of cancer types that had not yet been adequately tested for their response to lovastatin exposure. Our data showed an increased sensitivity to lovastatin-induced apoptosis in a number of tumor types, including head and neck squamous cell carcinomas (HNSCC) and cervical carcinomas (CC), at therapeutically achievable levels of this drug [6,9]. Based on these data, we believe that lovastatin may represent a novel therapeutic approach in HNSCC and CC and a Phase I trial evaluating the potential of lovastatin in the treatment of recurrent metastatic HNSCC and CC patients is currently underway at this Institute (Princess Margaret Hospital, Toronto, Ontario, Canada).
The mechanism and mediators of lovastatin-induced apoptosis of squamous cell carcinomas cells are currently unknown. Identification of these mediators may have clinical relevance by potentially uncovering predictors of response to this agent or novel therapeutic targets. In this study, we identified differentially expressed genes coincident with lovastatin treatment in HNSCC cell lines in an attempt to delineate the mechanism of its anticancer properties. Previous work, including our own, has clearly demonstrated that the identification of differentially expressed genes represents a feasible approach for elucidating mediators of the effects of various anticancer agents [3,5,7,10,20]. In this study, we used the Clontech Atlas Human Cancer Membrane Array spotted with 588 cancer-related cDNAs to identify lovastatin-regulated genes in two HNSCC-derived cell lines. We identified nine cDNAs that were modulated by lovastatin in both HNSCC cell lines, representing less than 2% of the cDNAs tested.
Because the biosynthesis of mevalonate leads to its incorporation into more than a dozen classes of end products that are vital for a variety of critical cellular functions [17], their role in lovastatin-induced apoptosis was also evaluated. Addition of the metabolites of all the major branches of the mevalonate pathway to determine which can modulate the apoptotic effects of lovastatin in HNSCC and CC cells may elucidate lovastatin mechanisms of action. We used this approach to identify potential mediators of the apoptotic effects of lovastatin in HNSCC and CC.
Materials and Methods
Tissue Culture
The HNSCC cell lines SCC9 and SCC25, the CC cell line SIHA, and the nontransformed monkey kidney cell line cos-7 were obtained from the American Tissue Culture Collection (Rockville, MD). The cell lines SCC9 and SCC25 were maintained in Dulbecco's MEM, and the SIHA and cos-7 cell lines in alpha-MEM (Princess Margaret Hospital Media Services) supplemented with 10% fetal bovine serum (Sigma, St. Louis, MO). The establishment of cos-7 cells constitutively expressing activated sterol response element-binding protein (SREBP) was performed by transfection using Effectene (Qiagen, Mississauga, Ontario, Canada) following the manufacturer's protocol. The expression plasmid containing the human SREBP-1a cDNA amino acids 1 to 487 or an empty vector control were provided and described by Dr. J. V. Swinnen (Catholic University of Leuven, Leuven, Belgium) [37]. Cells were exposed to solvent control or to 0 to 50 µM lovastatin (generously provided by Merck Research Laboratories, Rahway, NJ and diluted from a 10 mM stock in ethanol prepared as previously described [10]). The addback experiments utilized mevalonate, cholesterol, squalene, ubiquinone, farnesyl pyrophosphate (FPP), and geranylgeranyl pyrophosphate (GGPP) (all purchased from Sigma) as previously described [43].
cDNA Expression Microarray Blots
Total RNA was isolated from both SCC9 and SCC25 cell lines treated with solvent control or 10 µM lovastatin for 24 hours using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA were synthesized using the CDS primer mix (Clontech Laboratories, Palo Alto, CA) in the presence of 32P dCTP (final probe concentration of ∼0.5-1x106 cpm/ml) and hybridized to the Atlas Human Cancer Array (cat no. 7742-1; Clontech Laboratories) as per manufacturer's instructions and previously published reports [31,34]. After the posthybridization washes in a solution of SSC and SDS (2x SSC, 1% SDS)x3 (0.1 x SSX, 0.5% SDS)x2, the blots were exposed to Kodak Biomax MS X-ray film at x70°C for varying lengths of time to achieve optimal signal discrimination. The complete list of genes present on the Atlas Cancer Array can be obtained from Clontech Laboratories' web site (http://www.clontech.com/archive/APR98UPD/Atlaslist.html). The autoradiograms were digitized with the Agfa Studiostar scanner (Agfa, Toronto, Ontario, Canada). The Atlas Image software (Clontech Laboratories) was used to normalize the TIFF images of each expression array with respect to the levels of nine housekeeping genes provided on the blot. The background value was determined by measuring the mean basal signal intensity in regions of the autoradiogram away from DNA targets. Relative expression levels of solvent control versus lovastatin (10 µM for 24 hours)-treated cells were obtained by comparing intensities for each gene in the array and measuring the difference in signal intensity between them. The following expression values in units x103 of signal intensity were assigned for upregulated genes, as per Atlas Image software guidelines: “+” (range 14–20); “++” (21–27); “+++” (28–34); “++++” (35 and above). Any signal intensity close to background level was considered as no change. Minus symbols (“-”) denote downregulated expression using the same criteria as above. Only genes whose expression was altered in both the SCC9 and SCC25 cell lines were included as differentially expressed.
Luciferase Promoter Activity Assay
SCC9, SCC25, and cos-7 cells were seeded in 35-mm dishes at a density of 5x105 cells. The cells were incubated overnight to allow for cell attachment and recovery. The cell cultures were then transfected using 0.5 µg of the indicated luciferase plasmid construct and 0.5 µg of a β-galactosidase reporter construct using Effectene (Qiagen, Mississauga, Ontario, Canada) following the manufacturer's method. The diazepam-binding inhibitor (DBI), HMG-CoA reductase, and the HMG-CoA synthase promoter luciferase constructs were kindly provided by Dr. J. V. Swinnen (Catholic University of Leuven) [36,37]. The activated transcription factor (ATF) consensus luciferase construct was kindly provided by Dr. T. Hai (Ohio State University, Columbus, OH) [18,28]. The cells were then treated for 24 hours as indicated and harvested in 200 µl of reporter lysis buffer (Promega, Nepean, Ontario, Canada). Aliquots of 10 µl were assayed for luciferase activity using the luciferase assay reporter kit from Promega and a Turner 20/20 tube luminometer. The activity of β-galactosidase was used to normalize the transfection efficiencies.
MTT Assay
In a 96-well flat bottom plate (Nunc, Naperville, IL), approximately 5000 cells per 150 µl of cell suspension was used to seed each well. The cells were incubated overnight to allow for cell attachment and recovery. Following 2 days of treatment, 50 µl of 5 mg/ml MTT tetrazolium substrate (ICN, Toronto, Ontario, Canada) solution in phosphate-buffered saline was added and incubated for 6 hours at 37°C. The resulting violet formazan precipitate was solubilized by addition of 100 µl of a 0.01 M HCl/10% SDS (Sigma) solution shaking overnight at 37°C. The plates were then analyzed on an SLT Labinstruments 340 ATTC ELISA plate reader at 450 nm to determine the optical density of the samples.
Western Blot Analysis
Total cellular protein was extracted using a buffer that consisted of 1% NP40 (Sigma), 0.5% sodium deoxycholate (Sigma), 0.1% SDS (Sigma), 10 mg/ml leupeptin, and 2 trypsin inhibitory units/ml aprotinin (Sigma) in 2x PBS. Approximately 200 µl of extraction buffer was used to treat 106 cells. Total protein was quantified with the Biorad Protein Assay using bovine serum albumin (Sigma) as standard. Protein extracts representing 20 µg of total protein from the cell lines and their treatments were separated on a 10% SDS-PAGE gel and electrophoretically transferred onto PVDF membranes (Millipore, Toronto, Ontario, Canada). Membranes were blocked in 5% skim milk powder in PBS overnight at 40°C. Primary antibody, diluted in 10% FBS in PBS, was incubated with the membrane for 1 hour at room temperature. The antibodies used were specific for rhoA (Santa Cruz Biotechnology, Santa Cruz, CA) and actin (Sigma). The secondary antibodies (Amersham) were applied at a 1:5000 dilution in 3% BSA, 10% FBS in PBS, and incubated for 1 hour at room temperature [washes following antibody incubations are 3x5 minutes in PBS/0.05% Tween 80 (Sigma) then processed for chemiluminescent detection (Amersham)]. After the desired exposure was obtained, the membrane was stained with Coomassie Blue (Sigma) to ensure equal loading of the samples.
Phalloidin Staining
In a four-well Labtek Chamber slide (Nunc), approximately 10,000 cells in a 1-ml suspension was used to seed each well. The cells were incubated overnight to allow for cell attachment and recovery. Following 24 hours of treatment, the cells were washed twice with PBS and fixed for 5 minutes in 1.5% neutral buffered formalin (Sigma) at room temperature. The cells were then permeabilized using 0.1% Triton X-100 (Sigma) in PBS for 15 minutes. After two washes in PBS, the cells were incubated with FITC-conjugated phalloidin (Sigma) (1 µg/ml in PBS) for 15 minutes at room temperature. After two washes in PBS, the slides were mounted with a DAPI containing immunofluorescent mounting media (Vector Laboratories, San Diego, CA) and viewed by immunofluorescent microscopy.
Results
Microarray Analysis of Lovastatin Treatment in HNSCC Cell Lines
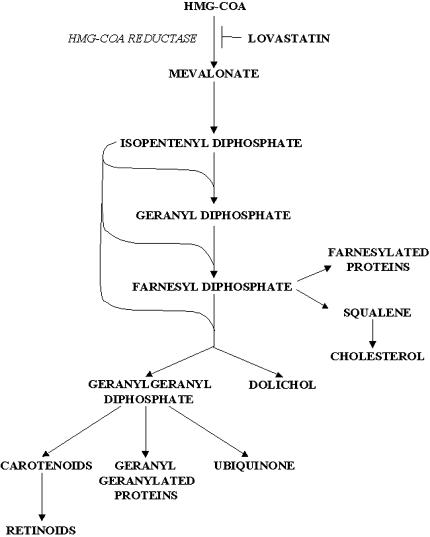
The cellular concentrations of mevalonate metabolites, which include de novo cholesterol, dolichol, ubiquinone, FPP, and GGPP (Figure 1), can affect the activity of various transcription factors [17]. For example, lower cholesterol levels can activate SREBP that binds sterol response elements (SRE) located in the promoters of various genes affecting their transcription [12,35]. The posttranslational modifications of a number of signaling proteins through the addition of the isoprenoid moieties farnesyl and geranylgeranyl are critical to their function, including their effects on transcriptional regulation [16,17,33]. Due to the potential of lovastatin to modulate transcription through the depletion of mevalonate products, we evaluated the effects of this drug on the expression of a wide range of genes using microarray analysis.
Figure 1.
The mevalonate pathway.
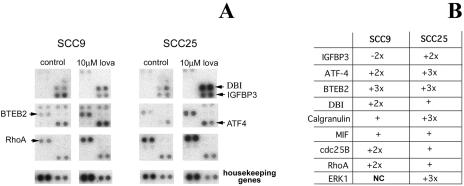
In this study, the HNSCC-derived cell lines SCC9 and SCC25 were treated with solvent control and 10 µM lovastatin for 24 hours. These conditions are prior to the onset of overt apoptosis that is triggered by lovastatin in these cell line models [9]. This response is dose-and timedependent and the above conditions were thus specifically used to identify potential triggers of lovastatin-induced apoptosis [9,10]. Total RNA was extracted from treated cells and relative expression data of 588 genes were generated using the Clontech Atlas array system [31,34] (Figure 2A). Of the 588 genes examined, only nine showed significant differences between control and lovastatin-treated cells (Figure 2B). These differences included genes involved in the ras signaling cascade [rhoA [2], extracellular signal-regulated kinase 1 (ERK1) [30]], growth regulation (insulin-like growth factor-binding protein 3 [21], cdc25B [11,26]), transcription factors [ATF-4 [18], basic transcriptional element-binding protein 2 (BTEB2) [42]], calcium regulation [calgranulin, migration inhibitory factor (MIF)-related protein [22,23]] and metabolism (DBI [36,37]). Of interest, DBI is a known SREBP-regulated gene [36,37] and the ATF family of transcription factors regulates the expression of a large number of genes involved in cell growth, differentiation, and homeostasis. Furthermore, a member of this family is also released from the endoplasmic reticulum by the same protease that cleaves SREBP [18]. This potential mechanistic link was further evaluated.
Figure 2.
(A) Differential expression analysis using the Clontech Atlas microarray between solvent control and treatment with 10 µM lovastatin in the SCC9 and SCC25 HNSCC-derived cell lines. (B) Differentially expressed genes were identified using the Atlas Image software. Of the 588 cDNAs represented on this slotblot, only nine demonstrated differential expression and are listed in the table with relative differences in expression (see Materials and Methods section for detailed description of analysis). The genes identified as potentially regulated by lovastatin included rhoA, extracellular signal-regulated kinase 1 (ERK1), insulin-like growth factor-binding protein 3 (IGFBP3), cdc25B, ATF-4, BTEB2, calgranulin, MIF-related protein, and DBI.
Promoter Analysis of DBI and the ATF Consensus Sequence
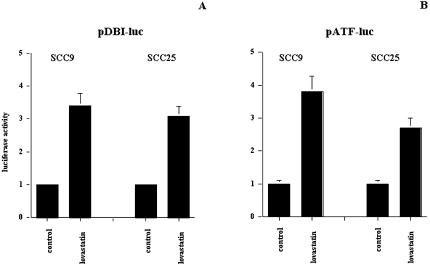
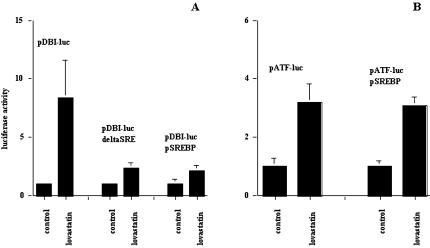
To evaluate the effects of lovastatin on DBI expression and the ability of lovastatin to regulate ATF target genes, we used a luciferase-based promoter assay. The DBI promoter [36,37] and a plasmid construct that contained the consensus-binding site for the ATF family of transcription factors were used [18,28]. Using both constructs, lovastatin induced three-to four-fold increases in expression from these promoters in both the SCC9 and SCC25 cell lines (Figure 3). These results confirm DBI as a target of lovastatin exposure, but also demonstrate that ATF target genes may be regulated and play a role in the biological effects of targeting HMG-CoA reductase in these cells.
Figure 3.
Promoter activity, as assessed by luciferase activity, of DBI and the ATF consensus constructs in SCC9 and SCC25. Cells were treated for 24 hours with solvent control or 10 µM lovastatin. Control levels of expression were normalized to one for ease of presentation.
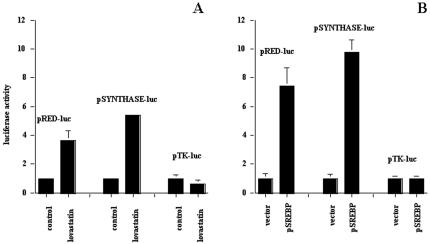
To further evaluate the effect of lovastatin on the expression of DBI and ATF target genes, we evaluated lovastatin's effects on these promoters in the nontransformed monkey kidney cell line cos-7. In this model, we confirmed that two mevalonate pathway enzymes that are known to be regulated by lovastatin through SREBP, HMGCoA synthase, and HMG-CoA reductase [37] show three- to six-fold induction with lovastatin, whereas the thymidine kinase (TK) promoter was not affected (Figure 4A). To establish the role of SREBP in these responses and to evaluate its potential role on the effects of lovastatin on the DBI promoter and the ATF consensus promoter constructs, we constitutively expressed the activated form of SREBP in the in the cos-7 cell line. As expected, the expression of activated SREBP resulted in elevated basal expression of both HMG-CoA synthase and HMG-CoA reductase, but the minimal TK promoter that lacks an SRE was unaffected compared to empty vector transfection control (Figure 4B). These results demonstrated that activated SREBP expressed in cos-7 cells was functional.
Figure 4.
(A) The effect of lovastatin on the expression of the HMG-CoA reductase (pRED-luc), the HMG-CoA synthase (pSYNTHASE-luc), and the thymidine kinase (pTK-luc) luciferase chimeric constructs in cos-7 cells. (B) Expression levels of pRED-luc, pSYNTHASE-luc, and pTK-luc in cos-7 cells constitutively expressing activated SREBP. Control levels of expression were normalized to one for ease of presentation.
For the DBI promoter, we evaluated the effects of lovastatin in cos-7 cells, with a DBI promoter construct that lacked its SRE and in cos-7 cells constitutively expressing active SREBP. The DBI promoter exhibited similar but more pronounced effects to the HNSCC cell lines with a 5- to 12-fold activation in response to treatment with 10 µM lovastatin for 24 hours (Figure 5A). Deletion of the SRE within this promoter construct abrogated this induction (Figure 5A). Overexpression of activated SREBP muted the induction elicited by lovastatin as well (Figure 5A), and taken together, these results indicate that SREBP was modulating the response of the DBI promoter to lovastatin treatment.
Figure 5.
(A) The effect of lovastatin on the expression of the DBI promoter, the DBI promoter with the SRE deleted, and the DBI promoter in cos-7 cells overexpressing activated SREBP. (B) The ATF consensus promoter luciferase chimeric construct in cos-7 cells and in cos-7 cells constitutively expressing activated SREBP. Cells were treated for 24 hours with solvent control or 10 µM lovastatin. Control levels of expression were normalized to one for ease of presentation.
For the ATF consensus promoter construct, we evaluated the effects of lovastatin in cos-7 cells and in cos-7 cells constitutively expressing active SREBP. The ATF consensus promoter in cos-7 cells showed similar results to the HNSCC cell lines with a three-to four-fold induction following lovastatin exposure (Figure 5B). The ATF consensus promoter, in contrast to the DBI promoter, does not possess an SRE and was not affected by SREBP overexpression (Figure 5B). These results indicate that the potential effects of lovastatin on the expression of ATF target genes are not dependent on SREBP activation but upon the activation of the ATF family of transcription factors.
Biochemical Analysis of Lovastatin-Induced Apoptosis
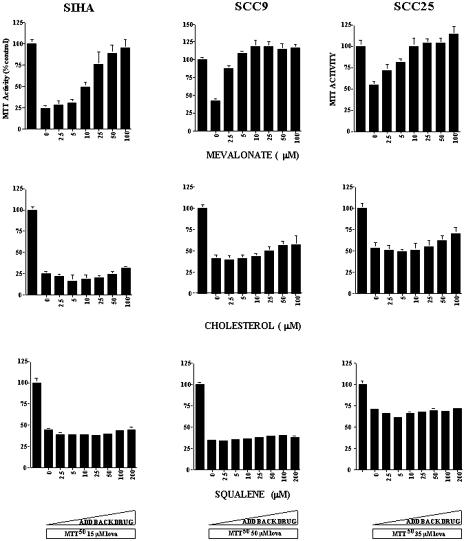
The cytotoxic effects of lovastatin and the inhibition of this cytotoxicity by the various products of the mevalonate pathway were evaluated using the MTT assay. We used the HNSCC cell lines SCC9 and SCC25 and the CC cell line SIHA. SIHA was included in this study as an alternative SCC derived from a different tissue site. The cell lines were treated with solvent control, a concentration of lovastatin that gave a 50% lethal dose [9], and a combination of lovastatin with increasing concentrations of the mevalonate metabolites used in this study. In all of the cell lines tested, the cytotoxicity triggered by the 50% lethal dose of lovastatin was inhibited following exposure to mevalonate in a dose-dependent manner (Figure 6). The concentration range tested was 2.5 to 100 µM and the inhibitory effect was evident at 10 µM concentration.
Figure 6.
The effect of adding back mevalonate, cholesterol, or squalene on the cytotoxic effects of lovastatin in the CC cell line SIHA and the HNSCC cell lines SCC9 and SCC25. Cell viability was assessed using the MTT assay. Cultures were treated with solvent control, lovastatin at its predetermined LD50 (SIHA 15 µM; SCC9 50 µM; SCC25 35 µM), and increasing concentrations of mevalonate (top row), cholesterol (middle row), and squalene (bottom row) for 48 hours.
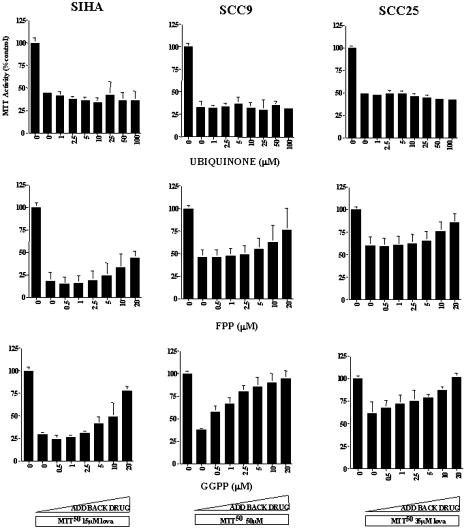
There are a number of end products downstream of mevalonate and it is, therefore, of interest to address which of these components is/are essential for lovastatin-induced cytotoxicity. In the cholesterol biosynthesis pathway, two products, squalene or cholesterol, were added to the culture medium at concentrations between 2.5 and 100 µM. These compounds were ineffective in blocking the cytotoxicity induced by lovastatin (Figure 6). The other mevalonate metabolite tested that had no protective effects on lovastatin-induced cytotoxicity was ubiquinone at concentrations of 1 to 100 µM (Figure 7).
Figure 7.
The effect of adding back ubiquinone, FPP, and GPP on the cytotoxic effects of lovastatin in the CC cell line SIHA and the HNSCC cell lines SCC9 and SCC25. Cell viability was assessed using the MTT assay. Cultures were treated with solvent control, lovastatin at its predetermined LD50 (SIHA 15 µM; SCC9 50 µM; SCC25 35 µM), and increasing concentrations of ubiquinone (top row), FPP (middle row), and GPP (bottom row) for 48 hours.
To determine whether protein isoprenylation is critical to the cytotoxic effects of lovastatin, the cell lines were also exposed to solvent control or a broad concentration range of GGPP or FPP. Coincubation with 0.5 to 20 µM GGPP blocked the loss of viability caused by lovastatin exposure in a dose-dependent manner. As shown in Figure 7, this protective effect was detectable at a range of 1.0 to 5.0 µM GGPP. Interestingly, at the same concentration range, FPP had only partial protective effects.
RhoA as a Potential Target of Lovastatin-Induced Apoptosis
In our microarray analysis, we identified rhoA as a gene that was potentially regulated by lovastatin. RhoA is a member of the Rho family of small GTPases that regulate actin cytoskeletal reorganization, thereby regulating cell shape and motility [2,14]. Rho proteins have been implicated in cancer cell invasion, growth, and survival [25,27], and require the isoprenoid moiety GGPP for their function. GGPP facilitates their membrane localization and transduction of upstream signals [27].
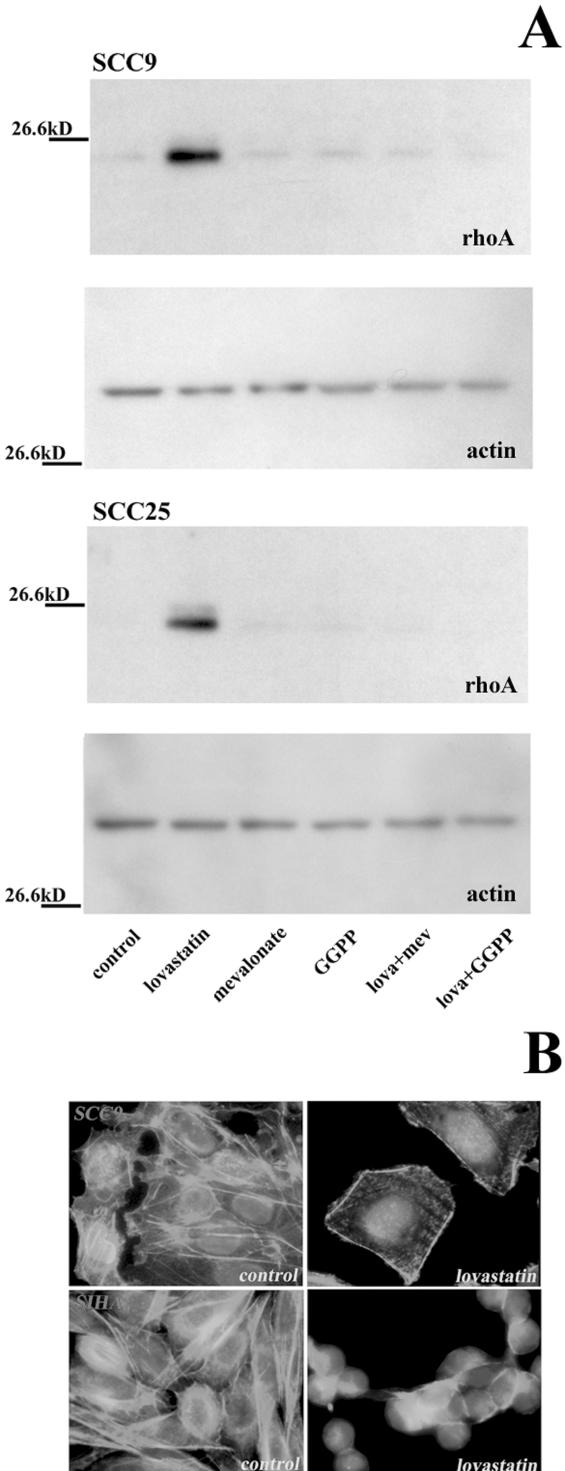
Using Western Blot analysis, we confirmed our microarray data for rhoA by demonstrating that protein levels were also upregulated in the SCC9 and SCC25 cell lines (Figure 8A). Furthermore, the addition of mevalonate and GGPP abrogated this response to lovastatin, indicating that the upregulation of rhoA in these SCC cells was the result of GGPP depletion following lovastatin exposure. Lovastatin did not affect the protein levels of actin in either of the cell lines tested using these conditions (Figure 8A). Due to the pivotal role of rhoA in regulating actin cytoskeletal organization, we also evaluated the effect of lovastatin on actin organization in treated cells. A fluorescent-conjugated phalloidin that possesses a strong binding affinity to actin was used to visualize actin's cytoskeletal organization. In untreated SCC9 and SIHA cells, actin stained in a filamentous pattern spanning the cell length (Figure 8B). Upon treatment with 10 µM lovastatin for 24 hours, this staining pattern was dramatically altered with distinct stress fibers forming on the periphery of the cells and disorganized actin in the cytoplasm. This pattern was observed prior to the induction of apoptosis as DAPI-counterstained nuclei did not display nuclear condensation or fragmentation characteristic of apoptosis (Figure 8B). These data verify that lovastatin's effect on rhoA expression and function occurred prior to the induction of apoptosis.
Figure 8.
(A) Western blot analysis of rhoA and actin in SCC9 and SCC25 cells treated for 24 hours with either solvent control, 10 µM lovastatin, 100 µM mevalonate, 10 µM GGPP, and combinations of lovastatin and mevalonate or GGPP. (B) Immunofluorescence microscopy to examine the effect of lovastatin on actin cytoskeletal organization in SCC cells. FITC phalloidin staining of SCC9 and SIHA cells with solvent control or 10 µM lovastatin for 24 hours. Nuclei were counterstained with DAPI. Original magnification, x400.
Discussion
The conversion of HMG-CoA to mevalonate is catalyzed by HMG-CoA reductase, the rate-limiting enzyme of the mevalonate pathway [17]. Mevalonate is the precursor of isoprene units incorporated into both sterol and nonsterol compounds such as cholesterol, dolichol, ubiquinone, GGPP, and FPP [17]. Cholesterol is a component of cellular membrane structure as well as a precursor for biosynthesis of steroid hormones and bile acids [17]. Dolichol, in its phosphorylated form, works as a carrier molecule of oligosaccharides in N-linked protein glycosylation [17]. Ubiquinone functions as an electron acceptor in the mitochondrial respiratory chain as well as an antioxidant with an important function in the inhibition of lipid peroxidation [17]. Farnesyl transferase and geranylgeranyl transferases utilize FPP and GGPP, respectively, for posttranslational isoprenylation of proteins [16,33]. The numbers of proteins that are modified by prenylation include ras and many small G proteins such as members of the Rab, Rac, and Rho families [16,32,33,38].
Inhibition of the mevalonate pathway by HMG-CoA reductase inhibitors results in depletion of mevalonate, the direct product of the enzyme reaction, and prevents the biosynthesis of downstream products inhibiting sterol synthesis, protein isoprenylation, and disruption of N-glycosylation [4,19]. A number of studies, including our own, have demonstrated that the targeting of HMG-CoA reductase leads to a pronounced tumor-specific apoptotic response that may represent a novel therapeutic approach in these cancers [1,6,9,10,29]. The mechanism by which depletion of these end products can result in a tumor-specific apoptotic response remains unknown. Because the cellular concentration of many of these products, in particular cholesterol, can affect the transcription of a number of genes by regulating the activity of various transcription factors [12,16,17], the modulators of the anticancer effects of lovastatin may be mediated by its gene targets. As mevalonate metabolites are involved in respiration, glycosylation, and posttranslational modification of proteins [16,17], the end products of this pathway that may be critical for the apoptotic effects of lovastatin may not involve direct transcriptional regulation of lovastatin-regulated genes.
We used microarray analysis to identify gene expression changes as potential mediators of lovastatin's apoptotic effects. We identified a number of genes that were regulated by lovastatin and further characterized two promoters that were regulated by sterol (DBI) and nonsterol (ATF consensus) mechanisms and the GGPP-modified small GTPase rhoA. The ATF family of transcription factors, which includes ATF-4, binds to the ATF consensus motif TGACGTCA that is present in many cellular promoters [18]. This suggests that this family of transcription factors may be involved in the regulation of the expression of multiple genes. To our knowledge, this is the first demonstration that targeting HMG-CoA reductase can potentially result in the regulation of ATF-driven promoters. The ability of lovastatin to activate the ATF consensus promoter through a nonsterol mechanism also suggests that its apoptotic effects may be mediated by nonsterol mevalonate metabolites. Our work indicates that this agent may activate a number of transcriptional regulatory pathways and validates this approach as a feasible method to identify mediators of lovastatin's anticancer effects.
We also evaluated the effect of various mevalonate metabolites on lovastatin's activity. Several studies have shown that mevalonate can rescue lovastatin-induced cell death in a variety of different tumor cells, including acute myeloid leukemias and medulloblastoma cells [41,43]. We found that mevalonate inhibited lovastatin-induced cell death in HNSCC and CC cell lines. Taken together, these results demonstrate that the cytotoxic effects of lovastatin are due to its ability to inhibit mevalonate synthesis. Our data clearly show that the depletion of mevalonate is responsible for lovastatin-induced apoptosis in HNSCC and CC cells, suggesting that blocking the production of specific mevalonate derivatives must be involved in this process. Cholesterol biosynthesis is the major metabolic branch in the mevalonate pathway and certain aspects of cholesterol metabolism appear to be relevant to cancer [17]. However, no protective effects against lovastatin cytotoxicity could be observed from the addition of squalene or cholesterol. These results suggest that the inhibition of cholesterol production is not critical to lovastatin-induced cytotoxicity in these cells.
In addition to cholesterol and squalene, we have also shown that ubiquinone is ineffective in preventing lovastatin-induced cytotoxicity in HNSCC and CC cells. It has been reported that ubiquinone supplementation prevents lovastatin -induced myopathy [39]; therefore, it may be a useful supplement for those patients unable to tolerate this side effect associated with lovastatin. The failure of these various compounds to overcome the cytotoxicity of lovastatin strongly suggests that other product(s) of mevalonate are involved in the control of cell survival. Prenylated proteins are posttranslationally modified at or near the carboxyl terminus by formation of cysteine thioethers with the isoprenoid lipid substrates FPP or GGPP [33]. These include small GTP-binding proteins, such as Ras, Rho, Raf, Rab, Rac, and Rap, that are involved in important cellular functions, such as the regulation of proliferation, signal transduction, intracellular transport, and cell death [32,38]. In this the study, we have shown that GGPP inhibited lovastatin-induced cytotoxicity in HNSCC and CC cells, whereas FPP only partially blocked lovastatin's effect.
The reversal of lovastatin-induced apoptosis in HNSCC and CC cells by GGPP is likely due to the replenishment of the intracellular pool of GGPP that is depleted by lovastatin treatment. By contrast, FPP leads only to a partial reversal of lovastatin-induced apoptosis and is likely the result of partial conversion of FPP to GGPP, as FPP is a substrate for other downstream metabolites including GGPP [17]. Alternatively, proteins that are normally geranylgeranylated may be farnesylated under conditions of intracellular GGPP shortage [27]. A number of studies have shown that GGPP can reverse the apoptotic effects of lovastatin in various tumor-derived cells including acute myeloid leukemias, colon adenocarcinomas, and medulloblastomas [1,8,29,41,43]. Our results imply a common mechanism or targets within these tumor types and HNSCC and CC. Furthermore, we identified rhoA as a lovastatin-regulated gene in SCC that requires GGPP modification for its membrane localization and function [2,27]. RhoA plays an integral role in actin cytoskeletal organization and regulates cell adhesion, morphology, motility, and invasion [2,14,25,27]. As such, rhoA has been implicated as a potential regulator of tumor cell invasion, growth, and survival [2,14,25,27]. Thus, rhoA may play a significant role in the anticancer properties of lovastatin. The identification of nonsterol-mediated gene targets of lovastatin and the GGPP reversal of its apoptotic effects implicate the nonsterol metabolites of mevalonate as mediators of lovastatin-induced apoptosis in HNSCC and CC. Targeting the formation of these nonsterol metabolites or their cellular targets may, therefore, lead to more refined therapeutic approaches.
Acknowledgements
Support from the Charlie Conacher Research Fund (S.K.R.), The Princess Margaret Hospital Foundation (S.K.R.), and the Canadian Institute of Health Research (fellowship; J.D.) is gratefully acknowledged. We also thank J. V. Swinnen and T. Hai for generously providing reagents used in this study.
Abbreviations
- HMG-CoA
3-hydroxy-3-methylglutaryl coenzyme A
- HNSCC
head and neck squamous cell carcinomas
- CC
cervical carcinomas
- SREBP
sterol response element-binding protein
- DBI
diazepam-binding inhibitor
- ATF
activated transcription factor
- GGPP
geranylgeranyl pyrophosphate
- FPP
farnesyl pyrophosphate
References
- 1.Agarwal B, Rao CV, Bhendwal S, Ramey WR, Shirin H, Reddy BS, Holt PR. Lovastatin augments sulindac-induced apoptosis in colon cancer cells and potentiates chemopreventive effect of sulindac. Proc Am Assoc Cancer Res. 1999;40:57. doi: 10.1016/s0016-5085(99)70342-2. [DOI] [PubMed] [Google Scholar]
- 2.Aznar S, Lacal JC. Rho signals to cell growth and apoptosis. Cancer Lett. 2001;165:1–10. doi: 10.1016/s0304-3835(01)00412-8. [DOI] [PubMed] [Google Scholar]
- 3.Cooper CS. Applications of microarray technology in breast cancer research. Breast Cancer Res. 2001;3:158–175. doi: 10.1186/bcr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corsini A, Maggi FM, Catapano AL. Pharmacology of competitive inhibitors of HMG-CoA reductase. Pharmacol Res. 1995;31:9–27. doi: 10.1016/1043-6618(95)80042-5. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham MJ. Genomics and proteomics: the new millennium of drug discovery and development. J Pharmacol Toxicol Methods. 2000;44:291–300. doi: 10.1016/s1056-8719(00)00111-8. [DOI] [PubMed] [Google Scholar]
- 6.Dimitroulakos J, Nohynek D, Backway KL, Hedley DW, Yeger H, Freedman MH, Minden MD, Penn LZ. Increased sensitivity of acute myeloid leukemias to lovastatin-induced apoptosis: a potential therapeutic approach. Blood. 1999;93:1308–1318. [PubMed] [Google Scholar]
- 7.Dimitroulakos J, Pienkowska M, Sun P, Farooq S, Zielenska M, Squire JA, Yeger H. Identification of a novel zinc finger gene, zf5-3, as a potential mediator of neuroblastoma differentiation. Int J Cancer. 1999;81:970–978. doi: 10.1002/(sici)1097-0215(19990611)81:6<970::aid-ijc21>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 8.Dimitroulakos J, Thai S, Wasfy GH, Hedley DW, Minden MD, Penn LZ. Lovastatin induces a pronounced differentiation response in acute myeloid leukemias. Leuk Lymphoma Res. 2000;40:167–178. doi: 10.3109/10428190009054894. [DOI] [PubMed] [Google Scholar]
- 9.Dimitroulakos J, Ye LY, Benzaquen M, Moore MJ, Kamel-Reid S, Freedman MH, Yeger H, Penn LZ. Differential sensitivity of various pediatric cancers and squamous cell carcinomas to lovastatin-induced apoptosis: therapeutic implications. Clin Cancer Res. 2001;7:158–167. [PubMed] [Google Scholar]
- 10.Dimitroulakos J, Yeger H. HMG-CoA reductase mediates the biological effects of retinoic acid on human neuroblastoma cells: lovastatin specifically targets P-glycoprotein-expressing cells. Nat Med. 1996;2:326–333. doi: 10.1038/nm0396-326. [DOI] [PubMed] [Google Scholar]
- 11.Eckstein JW. Cdc25 as a potential target of anticancer agents. Invest New Drugs. 2000;18:149–156. doi: 10.1023/a:1006377913494. [DOI] [PubMed] [Google Scholar]
- 12.Edwards PA, Tabor D, Kast HR, Venkateswaran A. Regulation of gene expression by SREBP and SCAP. Biochim Biophys Acta. 2000;1529:103–113. doi: 10.1016/s1388-1981(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 13.Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 14.Evers EE, Zondag GC, Malliri A, Price LS, ten Klooster JP, van der Kammen RA, Collard JG. Rho family proteins in cell adhesion and cell migration. Eur J Cancer. 2000;36:1269–1274. doi: 10.1016/s0959-8049(00)00091-5. [DOI] [PubMed] [Google Scholar]
- 15.Fisher DE. Apoptosis in cancer therapy: crossing the threshold. Cell. 1994;78:539–542. doi: 10.1016/0092-8674(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs JB. Ras C-terminal processing enzymes — new drug targets? Cell. 1991;65:1–4. doi: 10.1016/0092-8674(91)90352-y. [DOI] [PubMed] [Google Scholar]
- 17.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 18.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 19.Hunninghake DB. HMG-CoA reductase inhibitors. Curr Opin Lipidol. 1992;3:22–28. doi: 10.1097/00041433-199402000-00010. [DOI] [PubMed] [Google Scholar]
- 20.Kallioniemi OP. Biochip technologies in cancer research. Ann Med. 2001;33:142–147. doi: 10.3109/07853890109002069. [DOI] [PubMed] [Google Scholar]
- 21.Kelley KM, Oh Y, Gargosky SE, Gucev Z, Matsumoto T, Hwa V, Ng L, Simpson DM, Rosenfeld RG. Insulin-like growth factor-binding proteins (IGFBPs) and their regulatory dynamics. Int J Biochem Cell Biol. 1996;28:619–637. doi: 10.1016/1357-2725(96)00005-2. [DOI] [PubMed] [Google Scholar]
- 22.Kerkhoff C, Eue I, Sorg C. The regulatory role of MRP8 (S100A8) and MRP14 (S100A9) in the transendothelial migration of human leukocytes. Pathobiology. 1999;67:230–232. doi: 10.1159/000028098. [DOI] [PubMed] [Google Scholar]
- 23.Kerkhoff C, Klempt M, Kaever V, Sorg C. The two calcium-binding proteins, S100A8 and S100A9, are involved in the metabolism of arachidonic acid in human neutrophils. J Biol Chem. 1999;274:32672–32679. doi: 10.1074/jbc.274.46.32672. [DOI] [PubMed] [Google Scholar]
- 24.Kerr JF, Winterford CM, Harmon BV. Apoptosis. Its significance in cancer and cancer therapy. Cancer. 1994;73:2013–2026. doi: 10.1002/1097-0142(19940415)73:8<2013::aid-cncr2820730802>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 25.Kusama T, Mukai M, Iwasaki T, Tatsuta M, Matsumoto Y, Akedo H, Nakamura H. Inhibition of epidermal growth factor-induced RhoA translocation and invasion of human pancreatic cancer cells by 3-hydroxy-3-methylglutaryl-coenzyme a reductase inhibitors. Cancer Res. 2001;61:4885–4891. [PubMed] [Google Scholar]
- 26.Lammer C, Wagerer S, Saffrich R, Mertens D, Ansorge W, Hoffmann I. The cdc25B phosphatase is essential for the G2/M phase transition in human cells. J Cell Sci. 1998;111:2445–2453. doi: 10.1242/jcs.111.16.2445. [DOI] [PubMed] [Google Scholar]
- 27.Lebowitz PF, Prendergast GC. Non-Ras targets of farnesyltransferase inhibitors: focus on Rho. Oncogene. 1998;17:1439–1445. doi: 10.1038/sj.onc.1202175. [DOI] [PubMed] [Google Scholar]
- 28.Liang G, Hai T. Characterization of human activating transcription factor 4, a transcriptional activator that interacts with multiple domains of cAMP-responsive element-binding protein (CREB)-binding protein. J Biol Chem. 1997;272:24088–24095. doi: 10.1074/jbc.272.38.24088. [DOI] [PubMed] [Google Scholar]
- 29.Macaulay RJ, Wang W, Dimitroulakos J, Becker LE, Yeger H. Lovastatin-induced apoptosis of human medulloblastoma cell lines in vitro. J Neuro-Oncol. 1999;42:1–11. doi: 10.1023/a:1006164406202. [DOI] [PubMed] [Google Scholar]
- 30.Marais R, Marshall CJ. Control of the ERK MAP kinase cascade by Ras and Raf. Cancer Surv. 1996;27:101–125. [PubMed] [Google Scholar]
- 31.Pandita A, Zielenska M, Thorner P, Bayani J, Godbout R, Greenberg M, Squire JA. Application of comparative genomic hybridization, spectral karyotyping, and microarray analysis in the identification of subtype-specific patterns of genomic changes in rhabdomyosarcoma. Neoplasia. 1999;1:262–275. doi: 10.1038/sj.neo.7900036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruitt K, Der CJ. Ras and Rho regulation of the cell cycle and oncogenesis. Cancer Lett. 2001;171:1–10. doi: 10.1016/s0304-3835(01)00528-6. [DOI] [PubMed] [Google Scholar]
- 33.Sebti SM, Hamilton AD. Farnesyltransferase and geranylgeranyl transferase I inhibitors and cancer therapy: lessons from mechanism and bench-to-bedside translational studies. Oncogene. 2000;19:6584–6593. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 34.Sehgal A, Boynton AL, Young RF, Vermeulen SS, Yonemura KS, Kohler EP, Aldape HC, Simrell CR, Murphy GP. Application of the differential hybridization of Atlas Human expression arrays technique in the identification of differentially expressed genes in human glioblastoma multiforme tumor tissue. J Surg Oncol. 1998;67:234–241. doi: 10.1002/(sici)1096-9098(199804)67:4<234::aid-jso5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Shimano H, Yahagi N, Amemiya-Kudo M, Hasty AH, Osuga J, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Harada K, et al. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J Biol Chem. 1999;274:35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- 36.Swinnen JV, Alen P, Heyns W, Verhoeven G. Identification of diazepam-binding inhibitor/Acyl-CoA-binding protein as a sterol regulatory element-binding protein-responsive gene. J Biol Chem. 1998;273:19938–19944. doi: 10.1074/jbc.273.32.19938. [DOI] [PubMed] [Google Scholar]
- 37.Swinnen JV, Ulrix W, Heyns W, Verhoeven G. Coordinate regulation of lipogenic gene expression by androgens: evidence for a cascade mechanism involving sterol regulatory element binding proteins. Proc Natl Acad Sci USA. 1997;94:12975–12980. doi: 10.1073/pnas.94.24.12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 39.Thibault A, Samid D, Tompkins AC, Figg WD, Cooper MR, Hohl RJ, Trepel J, Liang B, Patronas N, Venzon DJ, et al. Phase 1 study of lovastatin, an inhibitor of the mevalonate pathway, in patients with cancer. Clin Cancer Res. 1996;2:483–491. [PubMed] [Google Scholar]
- 40.Thompson GR, Naoumova RP. Novel lipid-regulating drugs. Expert Opin Invest Drugs. 2000;9:2619–2628. doi: 10.1517/13543784.9.11.2619. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Macaulay RJ. Mevalonate prevents lovastatin-induced apoptosis in medulloblastoma cell lines. Can J Neurol Sci. 1999;26:305–310. doi: 10.1017/s0317167100000433. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe N, Kurabayashi M, Shimomura Y, Kawai-Kowase K, Hoshino Y, Manabe I, Watanabe M, Aikawa M, Kuro-o M, Suzuki T, et al. BTEB2, a Kruppel-like transcription factor, regulates expression of the SMemb/nonmuscle myosin heavy chain B, (SMemb/NMHC-B) gene. Circ Res. 1999;85:182–191. doi: 10.1161/01.res.85.2.182. [DOI] [PubMed] [Google Scholar]
- 43.Xia Z, Tan MM, Wong WW, Dimitroulakos J, Minden MD, Penn LZ. Blocking protein geranylgeranylation is essential for lovastatin-induced apoptosis of human acute myeloid leukemia cells. Leukemia. 2001;15:1398–1407. doi: 10.1038/sj.leu.2402196. [DOI] [PubMed] [Google Scholar]