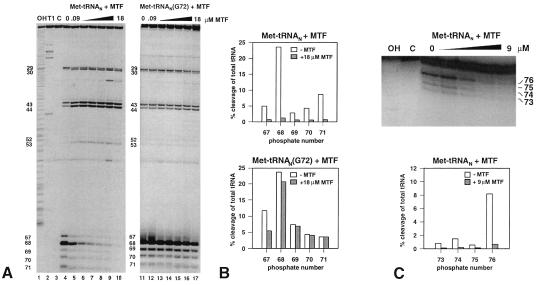
Figure 4.
(A) Electrophoretic analysis of partial RNase V1 cleavage of 3′-end [35S]Met-labeled initiator Met-tRNAN and Met-tRNAN (G72) on polyacrylamide gels. Met-tRNAN (lanes 4–10) and the G72 mutant Met-tRNAN (lanes 11–17) were incubated with increasing amounts of MTF (0, 0.09, 0.3, 0.9, 3, 9, and 18 μM) and subjected to partial digestion with RNase V1. Lane 1, OH, alkaline ladder; lane 2, T1, RNase T1 digest under denaturing conditions; lane 3, C, control without RNase VI treatment. (B) Quantitation of RNase VI cleavages in A by phosphorimager analysis. The amount of tRNA cut at a given position (67–71) is plotted as a percentage of total tRNA loaded per lane. Open bars, cleavage of Met-tRNAN, hatched bars, cleavage in the presence of 18 μM MTF. (C) Upper, electrophoretic analysis (on a 15% polyacrylamide gel) of a partial RNase T2 digest of 5′-end 32P-labeled Met-tRNAN. The tRNA was bound to increasing amounts of MTF (0.09–9 μM), and the complexes were analyzed by RNase T2 digestion; Lower, quantitation of RNase T2 footprinting by phosphorimager analysis.