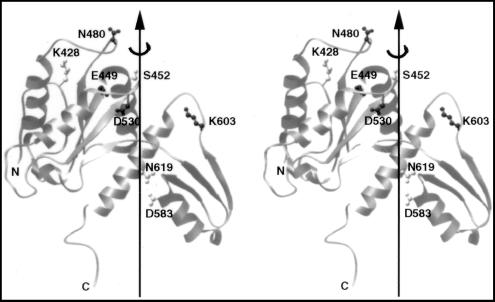
Figure 1.
A stereo view of the crystal structure of a fragment of yeast DNA topoisomerase II spanning amino acid residues 420 to 633. Eight of the ten residues selected for mutagenesis are shown in ball-and-stick models; the other two, Arg-650 and Lys-651, are within a region invisible in the published crystal structure of the yeast enzyme (8). Dark or light shading of a selected amino acid residue indicates its essentiality, as explained in the text. Drawing is based of the coordinates reported in ref. 8, by using the program ribbon (24).