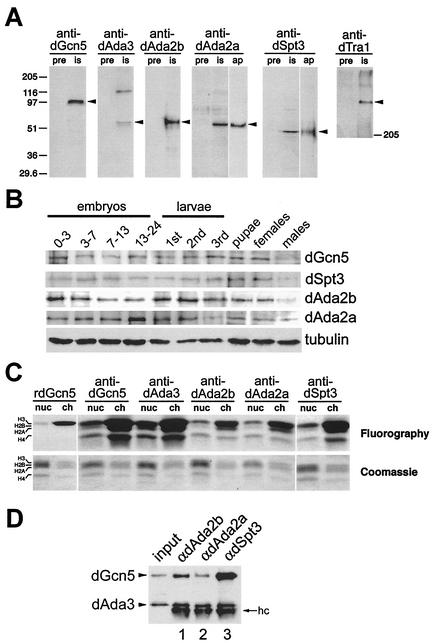
FIG. 2.
Characterization of the Drosophila GNAT subunits dAda2a, dAda2b, dAda3, dSpt3, and dTra1. (A) The antisera (is, immune serum) recognize proteins with the predicted molecular masses of dGcn5, dAda2b, dAda2a, dAda3, dSpt3, and dTra1 (arrowheads). Each lane contains 25 μg of nuclear extract. pre, preimmunization bleeds. Crude antisera against dAda2a and Spt3 detect a cross-reactive band of similar molecular mass, which is not recognized by affinity-purified antibodies (ap). (B) Developmental expression of dGcn5, dSpt3, dAda2a, and dAda2b. Lysates from Oregon R animals were electrophoresed on an SDS-10% polyacrylamide gel and analyzed by Western blotting. Embryonic lysates: 0-3 (blastoderm), 0-7 (gastrulation), 7-13 (germ band retraction), and 13-24 (differentiation). Larval stages: first, second, and third instars. Antibodies against tubulin served as loading controls. (C) HAT activity of the immunoprecipitates from 300 μg of S2 nuclear extract on nucleosomes (nuc) and core histones (ch), respectively. dAda3, dAda2a, dAda2b, and dSpt3 are associated with a dGcn5-specific HAT activity. Bacterially expressed rdGcn5 was used as a control. Top, fluorogram; bottom, photography of the Coomassie brilliant blue-stained SDS-polyacrylamide gel. (D) Western blot probed for dAda3 and dGcn5 (arrowheads). Input, 30 μg of nuclear extract; lane 1, immunoprecipitates from anti-dAda2b antibodies; lane 2, immunoprecipitates from anti-dAda2a; lane 3, immunoprecipitates from anti-dSpt3. hc, IgG heavy chain.