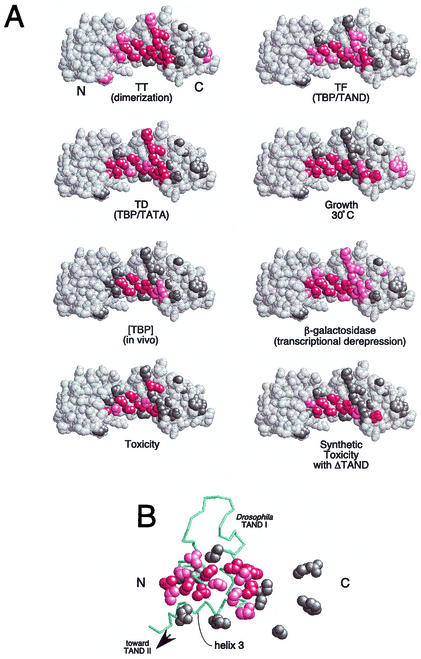
FIG. 4.
Summary of the properties of mutations along TBP's crystallographic dimer interface. (A) Shown are space-filling models of TBP monomers in the orientation shown in Fig. 1. N and C refer to the amino-terminal and carboxy-terminal stirrups, respectively. Each model is a summary derived from Table 1, in which each of the 24 tested amino acid side chains are color coded if mutations at these sites cause severe (red), moderate (pink), or no (gray) deviations from wild-type behavior. Since V71E but not V71R is defective for TAND binding (24), this residue was colored red. (B) The NMR structure of the Drosophila TAF1 TAND I backbone (from amino acid 19 to 77) is shown in the context of a space-filling representation of yeast TBP amino acid side chains that were used in this study (65). The color scheme is the same as that used in panel A. The view is that of panel A but rotated forward such that the TBP stirrups point inward and the convex seat of the saddle is facing outward.