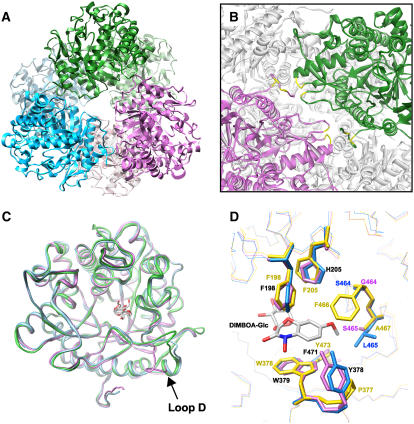
Figure 5.
Tertiary and quaternary structures of β-d-glucosidases. A, A ribbon diagram representation of the functional TaGlu1b hexamer. The dimers generated by crystallographic 2-fold symmetry operation are displayed as the same color. B, Side view of A (close-up view of dimer-dimer interface). The N-terminal regions of the monomer are positioned at the dimer-dimer interface. The four residues participating in direct hydrogen bonds to the monomer in the adjacent dimer are shown in yellow. These intermolecular hydrogen bonds are formed between T16 (side chain) and S366 (back bone), K17 (back bone) and Q270 (side chain), K19 (side chain) and S272 (side chain), and Q22 (side chain) and D271 (back bone). C, Superimposition of the back bones of TaGlu1b (magenta), ZmGlu1-E191D (PDB code 1E56; green), and SbDhr1 (PDB code 1V03; cyan). The structures of DIBOA-Glc and dhurrin in the substrate binding pockets of ZmGlu1 mutant and SbDhr1, respectively, are shown. D, Close-up view of the aglycone binding site. The crystal structures of TaGlu1b and ZmGlu1-E191D (PDB code 1E56) and the modeled structure of ScGlu are superimposed. A natural substrate, DIMBOA-Glc, bound to the maize enzyme is also shown. Blue, TaGlu1b; magenta, ScGlu; yellow, ZmGlu1-E191D. Identical residues found in TaGlu1b and ScGlu are shown in black. All of the molecular graphics images were produced using the UCSF Chimera package (Pettersen et al., 2004) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health P41 RR–01081).