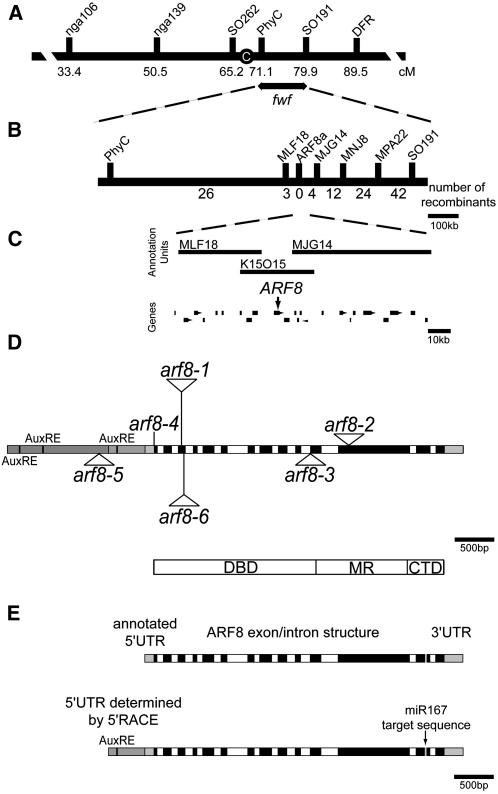
Figure 2.
Physical and Genetic Mapping of fwf.
(A) Genetic map of chromosome 5 showing genetic markers used in cloning fwf and the map position of fwf. C, centromere; cM, centiMorgan
(B) Physical map position of fwf with the number of recombinants between the markers indicated below.
(C) BAC vectors spanning the FWF region and the annotated genes delineated within this region. The position of ARF8 is indicated by the arrow.
(D) Diagram of the ARF8 gene with annotation of mutant alleles used in this study. T-DNA insertion lines are indicated by triangular insertions and the arf8-4 allele as a line, as it is an ethyl methanesulfonate mutation. Alleles and their ecotypes examined in this study were arf8-1 (Ws; Tian et al., 2004), arf8-4 (Ler; Vivian-Smith et al., 2001), arf8-5 (Col; Alonso et al., 2003), and arf8-6 (Col; http://www.hort.wisc.edu/krysan/DS-Lox/). Gray boxes indicate the promoter region and 3′ untranslated region (UTR). Three putative auxin response elements (AuxREs) are found in the promoter region. Black boxes specify exons, and white boxes indicate introns. A schematic representation of the ARF8 protein and its domains is represented below. DBD, DNA binding domain; MR, middle region; CTD, C-terminal domain.
(E) Structure of the ARF8 gene. Top: ARF8 gene with short 5′UTR as annotated in the database. Bottom: ARF8 gene with long 5′UTR as determined by 5′-RACE. The positions of the putative AuxRE present in the transcript leader region and of the microRNA target sequence (miR167) present in exon 13 are indicated.