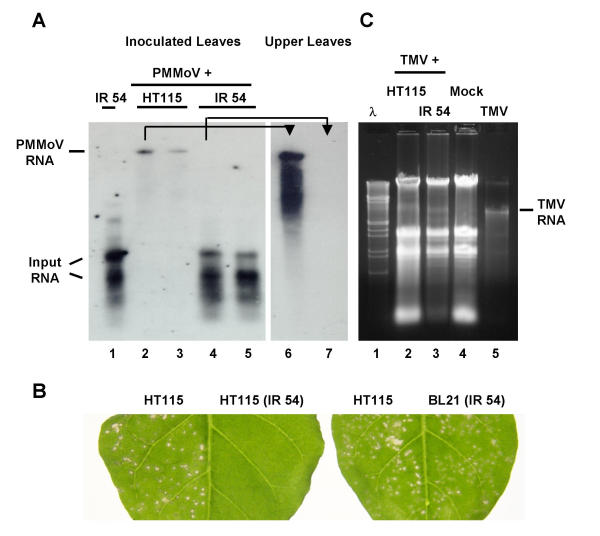
Figure 2.
HT115-expressed PMMoV IR 54 specifically interferes with PMMoV infection. (A) Northern blot analysis of total RNA extracted from inoculated (lanes 2 to 5) or uppermost systemic leaves (lanes 6 and 7) of N. benthamiana at 7 dpi. Plants were inoculated with mixtures of PMMoV (5 μg/ml) plus nucleic acid extracts prepared from HT115 harboring either pGEM/IR 54 (IR 54) or the empty vector (HT115). Extract from HT115/IR 54 used in the inoculum was run on lane 1 for comparison. RNA samples (1 μg) were fractionated by 1% agarose gel electrophoresis, and a DIG-labeled PMMoV 54-kDa RNA was used as a probe. (B) N. benthamiana plants were initially inoculated with mixtures of PMMoV plus bacterial nucleic acid extracts as indicated above. In addition, mixtures of PMMoV plus nucleic acid extracts prepared from BL21 carrying pGEM/IR 54 (BL21/IR 54) were included. After 7 days, 1:1000 diluted extracts from systemic leaves were assessed on opposite half-leaves of N. tabacum cv. Xanthi nc as indicated. Similar numbers of local lesions were observed in both halves of the leaf shown at the right. No visible local response was observed in the half-leaf inoculated with plant extracts derived from PMMoV plus HT115/IR 54 shown at the left. (C) Agarose gel analysis of total RNA (3 μg) extracted from systemic leaves of N. benthamiana plants that were mock inoculated or were inoculated with mixtures of TMV (5 μg/ml) plus nucleic acid extracts prepared from HT115 harboring either pGEM/IR 54 (IR 54) or the empty vector (HT115), as indicated. M, λEcoRI-HindIII molecular weight markers. TMV, purified TMV RNA (100 ng) was loaded as a control. The positions of PMMoV RNA, TMV RNA, and RNA species derived from partially denatured, input dsRNA are indicated.