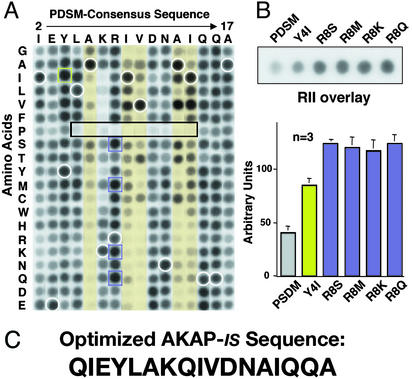
Figure 2.
Optimization of PDSM consensus sequence. (A) A two-dimensional array of 320 AKAP-IS peptide derivatives was generated where each residue between positions 2 and 17 in the PDSM sequence (above the array) was replaced with residues having every possible side chain (left of the array). Amino acids are indicated by their single-letter codes. Binding of 32P-labeled RII was detected by autoradiography. Peptide derivatives with substitutions at positions 6, 9, 10, 13, and 14 (yellow columns), proline substitutions with reduced RII binding (black rectangle), and internal control peptides of native sequence (white circles) are indicated. AKAP-IS derivatives with higher apparent RII-binding affinity (green and purple squares) are indicated. (B Upper) Solid-phase RII binding of the original PDSM consensus and the five peptides with higher affinity was quantified by densitometry of autoradiographs. Representative data from three individual experiments are presented. (B Lower) The relative binding affinity (arbitrary units) of each peptide is presented in graphic form. (C) The optimized AKAP-IS sequence.