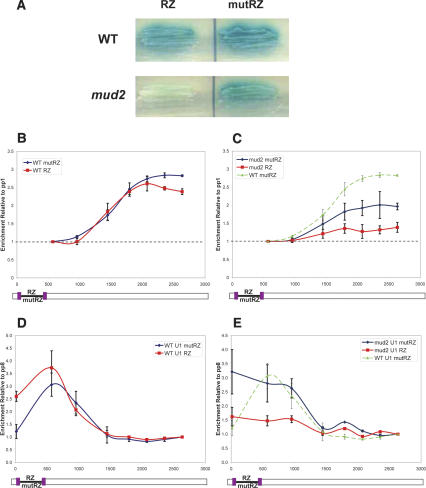
Figure 3.
Deletion of MUD2 sensitizes an intron to RZ cleavage. (A) Split MS2 derivatives of HZ18 with the RZ or mutRZ were transformed into wild-type (top panels) or Δmud2 (bottom panels) strains and assayed for β-gal activity as described (see Materials and Methods). Δmud2 increases the difference in blue intensity between the RZ and mutRZ reporters (bottom panels) compared to wild-type (top panels). (B,C) HA-MS2 ChIPs were performed as described (see Materials and Methods), and the data are presented as described in the legend to Figure 1. (B) MS2 ChIPs for the split MS2 mutRZ (blue) and RZ (red) reporters show little to no difference in a wild-type background. (C) In the Δmud2 background the split MS2 mutRZ (blue) signal is reduced compared to the same experiment in the wild-type strain (green), and the Δmud2 split MS2 RZ (red) signal is even further reduced. (D,E) U1 snRNP ChIPs were performed as described (see Materials and Methods), and the data are presented as described in the legend to Figure 1, except that the Y-axis represents the enrichment of DNA relative to the background signal seen at the last primer pair. (D) In a wild-type strain, U1 snRNP recruitment to the split MS2 RZ (red) is very similar to that for the split MS2 mutRZ (blue) aside from a modest increase at the first primer pair. (E) U1 snRNP recruitment in the Δmud2 strain is altered for both the split MS2 mutRZ (blue) as well as the split MS2 RZ (red) compared to the wild-type split MS2 mutRZ (green), and there is indeed a RZ-dependent reduction (compare red to blue).