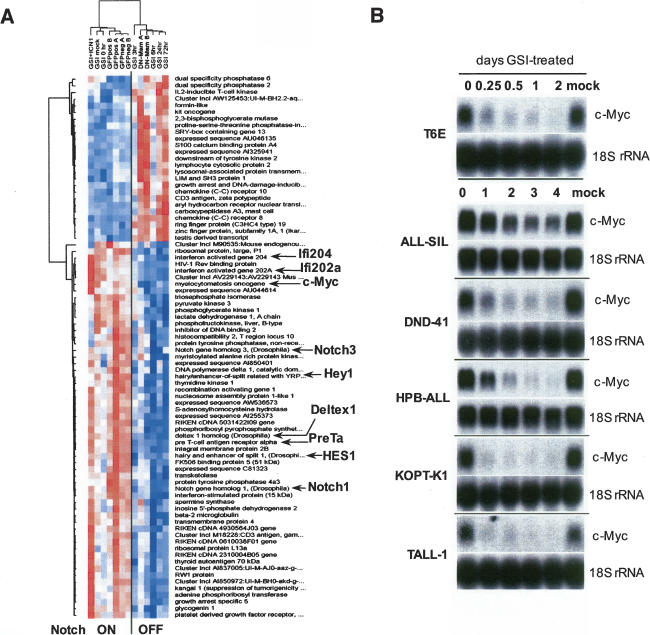
Figure 1.
Withdrawal of Notch1 signals down-regulates c-myc expression in T-ALL cells. (A) Identification of Notch1-sensitive genes in T6E cells. Columns represent experimental samples, while rows represent genes. Each colored box indicates relative expression level (normalized for each gene), where red indicates high and blue indicates low. The seven columns on the left are samples from T6E cells with active Notch signaling, and the six columns on the right are samples from T6E cells in which Notch signaling was inhibited. Expression data from 82 genes significantly correlated with the Notch “on” (left) versus “off” (right) distinction (p < 0.05) are depicted; expression of the 25 genes in the upper cluster increased on inhibition of Notch signaling, expression of the 57 in the lower cluster decreased. Genes implicated in the literature as related to Notch signaling as well as the novel gene c-myc are highlighted. Sample label key: (GSI [1 μM compound E] + ICN1) sorted ICN1 transduced cells treated with GSI; (GSI mock) DMSO vehicle-treated cells; (GSI 0 h) untreated cells; (GFPposA/B) sorted GFP-only cells; (GFPnegA/B) sorted untransduced cells from the same cultures as transduced cells; (GSI) cells treated with GSI for the indicated number of hours; (DN-MamA/B) sorted dominant-negative MAML1 transduced cells. (B) Northern blot analysis demonstrating the down-regulation of c-myc following treatment with GSI (1 μM compound E). Notch-dependent T6E cells and human T-ALL cell lines were treated with carrier (DMSO) or GSI for the indicated time periods. Blots were hybridized with probes indicated in the right margin.